
A novel genotype likelihood-based method to reduce mapping bias in low-coverage and ancient DNA studies
 based on reviews by Maxime Lefebvre, Michael Westbury and Adrien Oliva
based on reviews by Maxime Lefebvre, Michael Westbury and Adrien Oliva
Estimating allele frequencies, ancestry proportions and genotype likelihoods in the presence of mapping bias
Abstract
Recommendation: posted 08 March 2025, validated 13 March 2025
Ramos-Onsins, S. (2025) A novel genotype likelihood-based method to reduce mapping bias in low-coverage and ancient DNA studies. Peer Community in Genomics, 100410. 10.24072/pci.genomics.100410
Recommendation
The study of genomic variability within and between populations, as well as among species, relies on comparative analyses of homologous positions—sites that share a common evolutionary origin. Homology is inferred through sequence similarity (Reeck et al. 1987). However, the ability to detect homologous regions can be compromised when sequence mismatches accumulate due to mutations, especially when analyzing short DNA fragments, as in short-read sequencing (Li et al. 2008). In the genomic era, accurately mapping homologous DNA fragments to a reference genome is essential for obtaining precise estimates of genetic variability and evolutionary inferences (e.g., Li et al. 2008; Ellegren 2014). However, short-read, high-throughput sequencing often introduces mapping bias, disproportionately favoring the reference allele. This bias distorts allele frequency estimates, ancestry proportions, and genotype likelihoods, impacting downstream analyses (e.g., Günther & Nettelblad 2019; Martiniano et al. 2020).
Mapping bias is particularly problematic in ancient DNA studies, where post-mortem damage exacerbates sequencing errors. DNA fragmentation limits read length, while deamination, causing G to A and C to U transitions, increases mismatches and further complicates homology identification (Dabney & Pääbo 2013). These degradation processes contribute to the misidentification of true variants, confounding evolutionary inferences. Various strategies have been developed to mitigate mapping bias, including the commonly used approach, called pseudo-haploid data, that randomly picks a single read at each analyzed position for each individual, thereby retaining a single allele at each polymorphic site (Günther & Nettelblad 2019; Barlow et al. 2020).
Günther et al. (2025) introduce a novel method to correct mapping bias using a genotype likelihood-based approach, incorporating a mapping bias ratio to adjust for reference allele overrepresentation. The method specifically targets known single nucleotide polymorphisms (SNPs) because in population genomic analysis of ancient DNA data, low coverage and post-mortem damage often hinder the ability to identify novel SNPs in most individuals. The analysis focuses on DNA fragmentation, assuming that deamination effects are minimal when considering ascertained SNPs. The proposed method was compared against existing approaches, including pseudo-haploid data and standard genotype likelihood-based probabilistic methods. The evaluation was performed using both empirical and simulated data. For empirical data, low-coverage sequencing data from the 1000 Genomes Project (Finnish in Finland, Japanese in Tokyo, Yoruba in Ibadan, Nigeria populations) was analyzed, while for simulated data, ancient DNA-like datasets were generated using ms-prime (Kelleher et al. 2016), modeling different sequencing depths, divergence times, and reference genome choices. The study assesses the impact of mapping bias on the ratio of reference versus non-reference allele mapping, the accuracy of SNP allele frequency estimates relative to true frequencies, the deviation and variance between estimated and true allele frequencies, population differentiation and the estimation of admixture proportions using supervised and unsupervised methods, considering both genotype likelihoods and genotype calls.
Günther et al. (2025) bring to light that all methods analyzed exhibit minor but systematic reference allele bias. The new corrected genotype likelihood method outperforms the standard genotype likelihood approach in correlating with true allele frequencies, although the pseudo-haploid method still provides the most accurate estimates. Mapping bias also affects ancestry estimation, leading to admixture proportion errors of up to 4%, though this effect is smaller than the 10% discrepancy observed across different inference methods.
The work performed by Günther et al. (2025) provides a rigorous and innovative evaluation of mapping bias in the context of ascertained SNPs, introducing a probabilistic approach that improves bias correction. Unlike non-probabilistic methods such as pseudo-haploid data, the genotype likelihood framework leverages all sequencing reads for each analyzed SNP, and can incorporate additional bias corrections, enhancing its applicability across different sequencing conditions. While probabilistic approaches offer clear advantages in bias correction, they can be less intuitive to interpret compared to traditional genotype calling methods. This study highlights that mapping bias is pervasive across all methods, influencing evolutionary inferences such as selection signals and population differentiation. Although the improvements in allele frequency recovery may seem modest, the genome-wide impact of mapping bias is significant, especially in ancient DNA studies, making bias correction essential for robust evolutionary analyses.
References
Barlow A, Hartmann S, Gonzalez J, Hofreiter M, Paijmans JLA. (2020) Consensify: A method for generating pseudohaploid genome sequences from palaeogenomic datasets with reduced error rates. Genes;11(1):50. https://doi.org/10.3390/genes11010050
Dabney J, Meyer M, Pääbo S. (2013) Ancient DNA damage. Cold Spring Harb Perspect Biol. 5(7):a012567. https://doi.org/10.1101/cshperspect.a012567
Ellegren H. (2014) Genome sequencing and population genomics in non-model organisms. Trends Ecol Evol. 29(1):51-63. https://doi.org/10.1016/j.tree.2013.09.008
Günther T, Nettelblad C. (2019) The presence and impact of reference bias on population genomic studies of prehistoric human populations. PLoS Genet.15(7):e1008302. https://doi.org/10.1371/journal.pgen.1008302
Günther T., Goldberg A., Schraiber J. G. (2025) Estimating allele frequencies, ancestry proportions and genotype likelihoods in the presence of mapping bias. bioRxiv, ver. 5 peer-reviewed and recommended by PCI Genomics https://doi.org/10.1101/2024.07.01.601500
Kelleher J., Etheridge A. M., McVean G. (2016) Efficient coalescent simulation and genealogical analysis for large sample sizes. PLoS computational biology, 12(5):e1004842. https://doi.org/10.1371/journal.pcbi.1004842
Li H, Ruan J, Durbin R. (2008) Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18(11):1851-8. https://doi.org/10.1101/gr.078212.108
Reeck GR, de Haën C, Teller DC, Doolittle RF, Fitch WM, Dickerson RE, et al. (1987) "Homology" in proteins and nucleic acids: a terminology muddle and a way out of it. Cell. 50 (5): 667. https://doi.org/10.1016/0092-8674(87)90322-9
The recommender in charge of the evaluation of the article and the reviewers declared that they have no conflict of interest (as defined in the code of conduct of PCI) with the authors or with the content of the article. The authors declared that they comply with the PCI rule of having no financial conflicts of interest in relation to the content of the article.
Swedish Research Council Vetenskapsrådet (2017-05267) and Svenska Forskningsrådet Formas (2023-01381)
Evaluation round #3
DOI or URL of the preprint: https://doi.org/10.1101/2024.07.01.601500
Version of the preprint: 4
Author's Reply, 07 Mar 2025
Thank you for catching that missing reference and sorry for this omission! We have fixed the citation and added the reference to the reference list.
Decision by Sebastian Ernesto Ramos-Onsins , posted 07 Mar 2025, validated 07 Mar 2025
, posted 07 Mar 2025, validated 07 Mar 2025
Before recommendation please include the missing citation in the line 427 (?) and in the References section.
Evaluation round #2
DOI or URL of the preprint: https://doi.org/10.1101/2024.07.01.601500
Version of the preprint: 2
Author's Reply, 26 Feb 2025
Decision by Sebastian Ernesto Ramos-Onsins , posted 24 Feb 2025, validated 25 Feb 2025
, posted 24 Feb 2025, validated 25 Feb 2025
Dear Authors,
Thank you for submitting the revised version of your manuscript titled “Estimating Allele Frequencies, Ancestry Proportions, and Genotype Likelihoods in the Presence of Mapping Bias”. Both the reviewers and I are pleased with the improvements made in this version. The manuscript is now nearly ready for recommendation, pending a few minor revisions suggested by two of the reviewers.
Congratulations,
Reviewed by Maxime Lefebvre, 20 Feb 2025
Reviewed by Michael Westbury, 03 Feb 2025
Thank you for the detailed revision, it is much clearer what has been done now and it makes more sense.
I only have a few remaining comments.
Note the line numbers refer to the track changes document.
137: Typo + I thought the doGeno -4 was to not print genotypes?
193: Based on your reviewer response I think this refers to the haploid fasta mentioned above but I don't think the reader wouldn't know that just from stating "different reference genomes" here
251-253: While the short read lengths reflect aDNA, I think it is more likely the aDNA damage that influences mappability of the reads the most (as more mismatches mean less probablity of mapping) so I am not sure just shortening the reads produces results comparable to empirical aDNA damaged reads
Figure 3: What exactly is the Y-axis showing? I assume it is the different ancestry proportion but of which population, S2 or S3? Also, the dotted line across the figure is a little confusing as it only corresponds to one X-axis value
404-405: We found a similar result when mapping empirical data with simulated aDNA damage to different reference genomes and running Dstatistics with mapping to an ingroup producing the most reliable results - https://doi.org/10.1016/j.cub.2024.04.050 Supplementary figure S2 (Feel free not to cite it, I just thought it may be relevant)
Reviewed by Adrien Oliva, 20 Jan 2025
Thank you for addressing my comments and suggestions so thoroughly. I am pleased with the changes made to the manuscript, including the clarifications provided in your responses. The adjustments to the discussion, the inclusion of the JPT population, and the detailed explanation of your reproducibility measures significantly strengthen the paper.
I also appreciate the effort to correct typographical errors and improve the overall presentation of the manuscript. I am happy with the revisions and have no further comments. Thank you for your thoughtful work on this.
Evaluation round #1
DOI or URL of the preprint: https://doi.org/10.1101/2024.07.01.601500
Version of the preprint: 1
Author's Reply, 28 Dec 2024
Decision by Sebastian Ernesto Ramos-Onsins , posted 26 Sep 2024, validated 27 Sep 2024
, posted 26 Sep 2024, validated 27 Sep 2024
Dear Authors,
Thank you for submitting your study titled "Estimating allele frequencies, ancestry proportions and genotype likelihoods in the presence of mapping bias". It has been evaluated by three reviewers, all of whom recognize that your work offers interesting insights into the effects of mapping biases on frequency estimates and admixture proportion estimates, particularly in paleogenome studies that involve short-read sequences with relatively low read depth. While the reviewers acknowledge the improvements presented in your findings, they have also provided a number of comments, suggestions, and concerns to help enhance the manuscript.
A key concern raised by all three reviewers is the difficulty in reproducing your simulations and results. To address this, the authors must provide the complete code, with sufficient commentary, along with the specific options used in your pipelines, to ensure that the study can be fully replicated.
Additionally, the justification for focusing solely on ascertained biallelic sites in your study—particularly in the context of paleogenomics—needs to be strengthened. A more general comparative analysis that includes all sites, and contrasts with tools such as snpAD, might provide a more comprehensive understanding.
Since your approach seeks to mitigate mapping bias effects, it is also important to consider how varying mapping quality (MQ) values impact the analysis. In this study, MQ was fixed at 30, but assessing different MQ thresholds may provide additional insights into the robustness of your findings.
In the Results section, it would be beneficial to introduce clear subsections to distinguish between mapping biases in simulated data versus empirical data, and between the estimation of admixture proportions in these different contexts. The current structure may give the impression that empirical data and simulations are used arbitrarily across in analyses.
Finally, the Discussion section highlights that the improvement brought by your algorithm is relatively modest in comparison to other challenges in inference. As some reviewers have suggested, the Discussion would benefit from a more detailed exploration of the advantages of using this algorithm, including its computational cost, and the specific conditions or analyses where it proves most effective.
We look forward to receiving your revised manuscript.
Reviewed by Maxime Lefebvre, 06 Sep 2024
Reviewed by Michael Westbury, 30 Aug 2024
The manuscript by Gunther and Schraiber present a welcome and interesting addition to our knowledge of mapping/reference biases and how that can impact downstream analyses, especially relevant for palaeogenomic studies.
Overall I found it a sound study but lacking in some details in the methods that I think could help the reader understand what was done better and replicate it if needed.
Here are my specific comments that I hope will be helpful:
113-115: What are the original read lengths? 100bp or 150? Maybe even up to 300 if merged PE reads?
119: Why put this before the simulations? It seems strange to me to not give the details of the method first and just say "as described for the simulations below" since the reader has not read it yet. Normally I would put the details first and then write "see above"
123: What parameter does this in ANGSD? -anc ?
124-125: How was this done? and I don't quite understand what is meant by "the allele frequency estimation could be based on ANGSD"
127: Above it was past tense and now it is present tense. It would be good to keep it consistent
142: What does "according to the msprime simulations" mean?
142: What do you mean by first sequences?
144: gargammel uses a haploid fasta file to create the simulated reads. When/how was this sequence made? I assume you ran it independently for 2 individuals and merged the reads? A little more detail here would be helpful. Or did you directly use the msprime output and put it into gargammel? Since you mention this in the acknowledgments
147: What coverage was simulated?
150: This does seem on the low end as most studies use 30bp due to potential for spurious mapping e.g. https://doi.org/10.1093/bioinformatics/btae436. Could this impact the levels of reference bias?
151: Only merged reads were mapped?
155: How/when was genotype calling done? From the bam files?
156-158: I don't really understand what was done here
163: -GL 1 is using the SAMtools algorithm but above your said you tested the GATK one so I am a little confused. I know this is the pseudohaploid call but maybe it was also used for GL calling?
170: There is no mention how the GL were calculated? What tool was used?
170: An additional method that is gaining in popularity for ancestry estimation is admixfrog https://github.com/BenjaminPeter/admixfrog while only a suggestion it would be interesting to see how this performs relative to the other methods
171: But in the table there are only 4 shown
195: Why is the array less influenced by reference bias?
202: For someone who doesn't work with human data, what do these abbreviations stand for? Based on this I would assume YRI is more European as it shows more reference alleles?
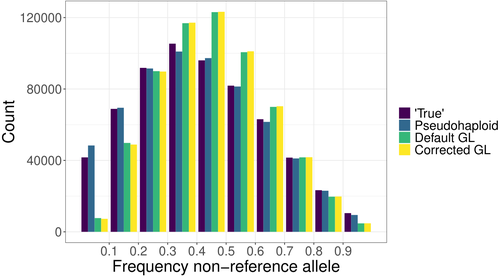
204: The phrasing here is a little difficult to interpret. Based on the plot it looks like GL finds the reference allele more relative to the "True" when the freq non-ref allele is low or high.
206: I am confused here. Looking at Fig2A and B the GL look the most different to the True in terms of counts whereas pseudohaploid looks more similar
213: I assume this means the correlations were done on a site by site basis? What could explain these differences between distribution and individual estimates?
220: what is a particular hint? a site?
300-302: Which specific fig/table does this refer to? I thought for GL there was a high correlation? I assume this refers to the outliers mentioned?
303-305: This is from Fig 2? if most non-reference alleles segregate at low frequency and you found when non-reference alleles are at a low frequency there is a lower count, doesn't this mean the opposite?
331: Where was the amount of missing data tested? I assume this is referring to the coverage of 0.5x and 2x?
In regards to the suggested list
Title and abstract
Does the title clearly reflect the content of the article? [x] Yes, [ ] No (please explain), [ ] I don't know
Does the abstract present the main findings of the study? [x] Yes, [ ] No (please explain), [ ] I don’t know
Introduction
Are the research questions/hypotheses/predictions clearly presented? [x] Yes, [ ] No (please explain), [ ] I don’t know
Does the introduction build on relevant research in the field? [x] Yes, [ ] No (please explain), [ ] I don’t know
Materials and methods
Are the methods and analyses sufficiently detailed to allow replication by other researchers? [ ] Yes, [x] No (please explain), [ ] I don’t know
Are the methods and statistical analyses appropriate and well described? [ ] Yes, [ ] No (please explain), [x] I don’t know
Results
In the case of negative results, is there a statistical power analysis (or an adequate Bayesian analysis or equivalence testing)? [ ] Yes, [ ] No (please explain), [x] I don’t know
Are the results described and interpreted correctly? [ ] Yes, [ ] No (please explain), [x] I don’t know
Discussion
Have the authors appropriately emphasized the strengths and limitations of their study/theory/methods/argument? [x] Yes, [ ] No (please explain), [ ] I don’t know
Are the conclusions adequately supported by the results (without overstating the implications of the findings)? [x] Yes, [ ] No (please explain), [ ] I don’t know
Reviewed by Adrien Oliva, 30 Aug 2024
Download the review









