
A new simulation pipeline enhances benchmarking of transposon polymorphism detection tools
Particular sequence characteristics induce bias in the detection of polymorphic transposable element insertions
Abstract
Recommendation: posted 12 May 2025, validated 21 May 2025
Castanera, R. (2025) A new simulation pipeline enhances benchmarking of transposon polymorphism detection tools. Peer Community in Genomics, 100418. https://doi.org/10.24072/pci.genomics.100418
Recommendation
Transposable Elements (TEs) are one of the main sources of genome variability. However, their study in populations has been hampered by the difficulty of properly detecting them using whole-genome re-sequencing data. Despite the expectations generated by the rise of long-read sequencing, today it is becoming clear that such technologies will not replace short-reads for analyzing large populations in the short term. Detecting Transposon Insertion Polymorphisms (TIPs) from short-read data is a challenging task, due to the repetitive nature of TE sequences that complicate read mapping. Nevertheless, accurate TIP detection is essential for understanding the evolutionary dynamics of TEs, their regulatory roles and their link with phenotypic variability. In the past 15 years, more than 20 tools have been developed for TIP detection using short-read data, but only a few independent benchmarks have been performed so far (Chen et al. 2023; Nelson et al. 2017; Rishishwar et al. 2017; Vendrell-Mir et al. 2019). Previous benchmarks have used simulated and real data to evaluate tool performance, each with its own set of advantages and limitations. In particular, introducing artificial insertions and simulating genomic short-reads may not reflect the nature of real TEs. By contrast, using real TE insertions as benchmarks can introduce bias since TE annotations are never perfect.
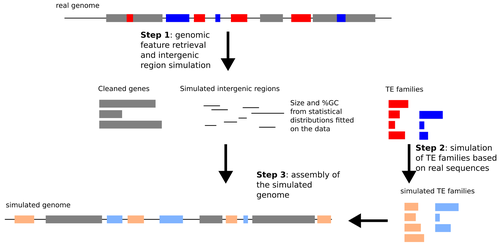
Verneret et al. (2025) introduce an original, alternative approach in which a comprehensive simulation method mimics the most important sequence features of real TEs and non-TE intergenic regions. This simulated data is then combined with true genic sequences, generating a pseudochromosome that can be used for benchmarking TIP detection pipelines. Using this approach, the authors eliminate the bias of TE annotation on real genomes, while preserving most of the characteristics of natural TEs. Using simulated pseudochromosomes for Drosophila melanogaster and Arabidopsis thaliana, Verneret et al. (2025) found that the performance of 14 commonly used TIP-calling tools is highly variable, with only a few performing well, and only at high sequencing depths. In addition to this, the authors analyzed the sequence features of true-positive and false-positive TIP calls, and found that specific TE sequence characteristics (e.g., length, age, etc.) affect the detection of both reference and non-reference TIPs.
The approach described by Verneret et al. (2025) is an important contribution to the field for several reasons. On the one hand, the results shown in the publication will help the users of such tools make informed decisions before launching their experiments. For more advanced users, it will enable future benchmarks to identify which tools perform best for different species, each with their own sequence characteristics. For software developers, the data released constitutes a precious dataset to test their tools in the same conditions. Finally, the identification of sequence characteristics enriched among false positives and false negatives also gives an opportunity for developers to improve the performance of the new tools by considering these specificities.
References
Chen J, Basting PJ, Han S, Garfinkel DJ, Bergman CM (2023) Reproducible evaluation of transposable element detectors with McClintock 2 guides accurate inference of Ty insertion patterns in yeast. Mobile DNA, 14, 8. https://doi.org/10.1186/s13100-023-00296-4
Nelson MG, Linheiro RS, Bergman CM (2017) McClintock: An integrated pipeline for detecting transposable element insertions in whole-genome shotgun sequencing data. G3: Genes, Genomes, Genetics, 7, 2763–2778. https://doi.org/10.1534/g3.117.043893
Rishishwar L, Mariño-Ramírez L, Jordan IK (2017) Benchmarking computational tools for polymorphic transposable element detection. Briefings in Bioinformatics, 18, 908–918. https://doi.org/10.1093/bib/bbw072
Vendrell-Mir P, Barteri F, Merenciano M, González J, Casacuberta JM, Castanera R (2019) A benchmark of transposon insertion detection tools using real data. Mobile DNA, 10, 53. https://doi.org/10.1186/s13100-019-0197-9
Verneret M, Le VA, Faraut T, Turpin J, Lerat E (2025) Particular sequence characteristics induce bias in the detection of polymorphic transposable element insertions. bioRxiv, ver. 4 peer-reviewed and recommended by PCI Genomics https://doi.org/10.1101/2024.09.25.614865
The recommender in charge of the evaluation of the article and the reviewers declared that they have no conflict of interest (as defined in the code of conduct of PCI) with the authors or with the content of the article. The authors declared that they comply with the PCI rule of having no financial conflicts of interest in relation to the content of the article.
INRAE GoatRetrovirome grant , ANR-22-CE35-0002-01
Evaluation round #2
DOI or URL of the preprint: https://doi.org/10.1101/2024.09.25.614865
Version of the preprint: 3
Author's Reply, 30 Apr 2025
Dear Raúl,
Thank you again for this opportunity to improve our manuscript. We have taken into account the various remarks made by the reviewer as you will see in our answers, in the text and with the additional files we provide. We hope that you will find them suitable.
Best,
Marie Verneret and colleagues
Reviewer 1
· Materials and methods
Are the methods and analyses sufficiently detailed to allow replication by other researchers? No
The description, code and documentation for generating simulated genomes are described well, but several key aspects of the methodology are not detailed that prevent replication by other researchers.
1) There is no code or documentation for how the TE detection tools were run
Answer: we now provide as supplementary data (Supplementary file 1) the command lines used to run TEPID and Jitterbug. Regarding the other programs, they were run via McClintock2 so there is no particular code, just the command line as indicated in the McClintock2 github.
2) There is no documentation for running the code to perform the evaluation
Answer: we have put on the github of replicaTE a readme page to indicate how to run the different programs used for the evaluation. Moreover, we provide as supplementary data (Supplementary file 2) the various pre-processing steps before running the script FP_TP_teflon_insertion.py.
3) A description of the alignment methods used to project TE annotations for simulated genomes onto reference genome coordinates is not given
Answer: we have now added the names of the various alignment tools used and TE annotations are provided as supplementary data (Supplementary file 3).
4) long-read sequencing data and TE annotations for long-read dataset for Bos taurus are not made available. Making all of these resources available is essential for journal publication.
Answer: we have added in the Supplementary Table S1 all the accession numbers for both long and short reads.
· Specific comments
- line 80: “generally” -> “often”
Answer: this has been corrected
- line 82-3: “Vendrell-Mir (Vendrell-Mir et al. 2019)” -> “Vendrell-Mir (2019)”
Answer: this has been corrected
- line 86: “Rishiwar et al. (Rishishwar et al. 2017)” -> “Rishiwar et al. (2017)”
Answer: this has been corrected
- line 88: “Nelson et al. and Chen et al. (Nelson et al. 2017; Chen et al. 2023)” -> “Nelson et al. (2017) and Chen et al. (2023)”
Answer: this has been corrected
- line 89: “the yeast” -> “yeast”
Answer: this has been corrected
- line 91: “Then” -> “Hence”
Answer: this has been corrected
- line 127: describe what “cleaned up from any TE” means
Answer: we have added a sentence to explain this: « any TE inserted inside the genes, given the annotation, are removed from the gene sequences »
- line 142: say “see results for description of files”
Answer: this has been added
- line 149-50: “Target Site Duplication (TSD)” -> “TSD”
Answer: this has been corrected
- line 179-82: move description of scripts used to calculate TP, etc after definition of these measurements (i.e., after line 192).
Answer: the mention was already present where the reviewer wants to move it but thus was repeated. We removed the description from lines 179-182.
- line 201: state that the strand of the TE prediction and annotation is not considered in the performance evaluation
Answer: we now state that the strand has not been taken into account for the evaluation. However, the annotation (meaning the correct name family) is indeed taken into account.
- line 211: provide accessions for long read data
Answer: the accession numbers for all long and short read data are in the Supplementary Table S1.
- line 212: provide supplemental file of TE annotations based on long and short read data.
Answer: we provide as supplementary data (Supplementary file 3) these TE annotations.
- line 251: “ligth" -> “light”
Answer: this has been corrected
- line 261: provide details of how TE annotations in simulated genomes are projected back onto reference genome coordinates (whole genome alignment?, alignment of flanking regions?)
Answer: as we already indicated in the text, since we have the coordinates of all the insertions and given that the tested tools provide the coordinates of the polymorphic insertions they predict, we can thus determine whether these predictions are correct (according to a margin of error in bp) given that the correct name of the TE family is also predicted. There is no need to perform alignment.
- line 290: “On Figure” -> “In Figure”
Answer: this has been corrected
- line 296: the observation that TEMP/TMEP2 predicts more reference TE insertions than other methods is expected since the way that McClintock reports reference insertions for these methods is different than other McClintock components. Instead of finding evidence for the presence of a reference TE insertions, TEMP/TEMP2 find evidence for the absence of reference insertion, then McClintock finds the complement of the set of “non-absent” reference annotation to generate a set of reference TE calls. This tends to inflate the number of reference TE calls for TEMP/TEMP2.
Answer. the reviewer is right. Thank you for the remark. We have added this information in the text.
- line 296: “Other programs find more” -> “Other programs find fewer”
Answer: this has been corrected
- line 301: Figure 3, right panel – the dark yellow line should be labeled TEMP2
Answer: this has been corrected
- line 305: Figure 3 legend should say that some lines are overlapping and can’t be seen.
Answer: this has been corrected
- line 347: 2x “answers” -> “predictions”
Answer: this has been corrected
- line 348 “all the TE insertions” -> “all the reference TE insertions”
Answer: this has been corrected
- line 366: “We have then” -> “We then” →
Answer: this has been corrected
- line 367: “inferior to” -> “less than”
Answer: this has been corrected
- line 371 and elsewhere: underscores are incorrect in “ngs-te-mapper2” (please check all occurrences)
Answer: we have corrected the name of the tool in the whole manuscript.
- Line 451: “all” -> “both”
Answer: this has been corrected
- line 477: explain why TEPID and Jitterbug are not used in this analysis
Answer: since TEPID and Jitterbug did not perform very well with the two other species and since they have to be run independently, we preferred not to use them and use the tools only present in McClintock2.
- line 514: “dataset” -> “datasets”
Answer: this has been corrected
- lines 554-5: “they need to be not too divergent from the consensus or reference TE used to identify them” -> “they need to be similar to the consensus or reference TE sequences used to identify them”
Answer: this has been corrected
- line 586: add Chen et al 2019 to prior work showing evidence that coverage impacts non-reference TE detection
Answer: we have added the reference.
- line 604: “It is to note that TEBreak” - > “We note that RelocaTE2”
Answer: this has been corrected
- line 611: state which “two tools” were analyzed
Answer: we now state to which tools we were referring to (popoolationTE2 and TIDAL).
- line 620: “Vendrell-Mir (Vendrell-Mir et al. 2019)” -> “Vendrell-Mir (2019)”
Answer: this has been corrected
- line 630: It is possibly worth mentioning an alternative strategy to using the intersection of multiple methods to strengthen conclusions based on non-optmal performance of short-read TE detectors. Some authors acknowledge the non-optimal performance and instead of filtering to use predictions supported by multiple methods, they test whether the overall biological conclusion of a study is robust to the choice of TE detector (ie. Manee et al 29850787).
Answer: we thank the reviewer for this thought. It is certainly of great interest to also consider evolutionary and biological meanings when determining which polymorphic insertions may be indeed true positives. We have added the reference to this work.
Decision by Raúl Castanera, posted 10 Apr 2025, validated 10 Apr 2025
Dear Marie Verneret and co-authors,
Thank you for providing a revised manuscript following reviewers’ comments. I think your study is an important contribution to the TE community, both for software developers aiming to improve TIP detection and for the users of these tools, who often struggle to identify the best strategy to analyze their data. Nevertheless, as pointed out by one reviewer, there are still some minor but neccessary improvements needed before I can recommend your manuscript. These relate to the description of the methodologies (exact code/parameters used to reproduce some of the analyses), as well as some text corrections.
Best regards,
Raúl
Reviewed by anonymous reviewer 1, 05 Apr 2025
· Title and abstract
Does the title clearly reflect the content of the article? Yes.
Does the abstract present the main findings of the study? Yes
· Introduction
Are the research questions/hypotheses/predictions clearly presented? Yes
Does the introduction build on relevant research in the field? Yes
· Materials and methods
Are the methods and analyses sufficiently detailed to allow replication by other researchers? No
The description, code and documentation for generating simulated genomes are described well, but several key aspects of the methodology are not detailed that prevent replication by other researchers.
1) There is no code or documentation for how the TE detection tools were run
2) There is no documentation for running the code to perform the evaluation
3) A description of the alignment methods used to project TE annotations for simulated genomes onto reference genome coordinates is not given
4) long-read sequencing data and TE annotations for long-read dataset for Bos taurus are not made available.
Making all of these resources available is essential for journal publication.
Are the methods and statistical analyses appropriate and well described? Yes
· Results
In the case of negative results, is there a statistical power analysis (or an adequate Bayesian analysis or equivalence testing)? Yes
Are the results described and interpreted correctly? Yes
· Discussion
Have the authors appropriately emphasized the strengths and limitations of their study/theory/methods/argument? Yes
Are the conclusions adequately supported by the results (without overstating the implications of the findings)? Yes
· Specific comments
- line 80: “generally” -> “often”
- line 82-3: “Vendrell-Mir (Vendrell-Mir et al. 2019)” -> “Vendrell-Mir (2019)”
- line 86: “Rishiwar et al. (Rishishwar et al. 2017)” -> “Rishiwar et al. (2017)”
- line 88: “Nelson et al. and Chen et al. (Nelson et al. 2017; Chen et al. 2023)” -> “Nelson et al. (2017) and Chen et al. (2023)”
- line 89: “the yeast” -> “yeast”
- line 91: “Then” -> “Hence”
- line 127: describe what “cleaned up from any TE” means
- line 142: say “see results for description of files”
- line 149-50: “Target Site Duplication (TSD)” -> “TSD”
- line 179-82: move description of scripts used to calculate TP, etc after definition of these measurements (i.e., after line 192).
- line 201: state that the strand of the TE prediction and annotation is not considered in the performance evaluation
- line 211: provide accessions for long read data
- line 212: provide supplemental file of TE annotations based on long and short read data.
- line 251: “ligth" -> “light”
- line 261: provide details of how TE annotations in simulated genomes are projected back onto reference genome coordinates (whole genome alignment?, alignment of flanking regions?)
- line 290: “On Figure” -> “In Figure”
- line 296: the observation that TEMP/TMEP2 predicts more reference TE insertions than other methods is expected since the way that McClintock reports reference insertions for these methods is different than other McClintock components. Instead of finding evidence for the presence of a reference TE insertions, TEMP/TEMP2 find evidence for the absence of reference insertion, then McClintock finds the complement of the set of “non-absent” reference annotation to generate a set of reference TE calls. This tends to inflate the number of reference TE calls for TEMP/TEMP2.
- line 296: “Other programs find more” -> “Other programs find fewer”
- line 301: Figure 3, right panel – the dark yellow line should be labeled TEMP2
- line 305: Figure 3 legend should say that some lines are overlapping and can’t be seen.
- line 347: 2x “answers” -> “predictions”
- line 348 “all the TE insertions” -> “all the reference TE insertions”
- line 366: “We have then” -> “We then” ->
- line 367: “inferior to” -> “less than”
- line 371 and elsewhere: underscores are incorrect in “ngs-te-mapper2” (please check all occurences)
- Line 451: “all” -> “both”
- line 477: explain why TEPID and Jitterbug are not used in this analysis
- line 514: “dataset” -> “datasets”
- lines 554-5: “they need to be not too divergent from the consensus or reference TE used to identify them” -> “they need to be similar to the consensus or reference TE sequences used to identify them”
- line 586: add Chen et al 2019 to prior work showing evidence that coverage impacts non-reference TE detection
- line 604: “It is to note that TEBreak” - > “We note that RelocaTE2”
- line 611: state which “two tools” were analyzed
- line 620: “Vendrell-Mir (Vendrell-Mir et al. 2019)” -> “Vendrell-Mir (2019)”
- line 630: It is possibly worth mentioning an alternative strategy to using the intersection of multiple methods to strengthen conclusions based on non-optmal performance of short-read TE detectors. Some authors acknowledge the non-optimal performance and instead of filtering to use predictions supported by multiple methods, they test whether the overall biological conclusion of a study is robust to the choice of TE detector (ie. Manee et al 29850787).
https://doi.org/10.24072/pci.genomics.100418.rev21Evaluation round #1
DOI or URL of the preprint: https://doi.org/10.1101/2024.09.25.614865
Version of the preprint: 2
Author's Reply, 21 Mar 2025
Please find in the enclosed files our response to comments as well as the tracked change main document where all modifications are in red.
Decision by Raúl Castanera, posted 14 Nov 2024, validated 14 Nov 2024
Dear Marie Verneret and co-authors,
Your manuscript has been peer reviewed by two experts in the field. The two reviewers and I agree that your simulation tool and benchmark of TIP detection tools can be a valuable contribution for the TE community. Nevertheless, they have identified room for improvement in the description of the methodoloy and the integration of your results in the context of previous work. Also, some aspects of the simulation strategy have been critizied. They recommend a number of revisions that can enhance the quality of the work. I will be happy to consider a revised version of the manuscript for reccomendation.
Best wishes,
Raúl Castanera
Reviewed by anonymous reviewer 1, 28 Oct 2024
Title and abstract
- Does the title clearly reflect the content of the article? [ ] Yes, [ ] No (please explain), [x] I don't know
The title only reflects one of the main results from the the paper.
- Does the abstract present the main findings of the study? [x] Yes, [ ] No (please explain), [ ] I don’t know
Introduction
- Are the research questions/hypotheses/predictions clearly presented? [x] Yes, [ ] No (please explain), [ ] I don’t know
- Does the introduction build on relevant research in the field? [ ] Yes, [x] No (please explain), [ ] I don’t know
Prior efforts developing simulation systems to evaluate the performance of TE detection systems are not described (i.e., simulaTE by Kofler 2018 and the single TE insertion framework by Nelson et al 2017/Chen et al 2023).
Materials and methods
- Are the methods and analyses sufficiently detailed to allow replication by other researchers? [ ] Yes, [x] No (please explain), [ ] I don’t know
The version and parameters used to generate the simulated Drosophila, Arabidopsis and Bos genomes using replicaTE are not given. The version and parameters used to run the McClintock TE detection system, TEPID and Jitterbug are not given. The accessions for the data used in the analysis of Bos short and long read sequences are not provided. The version and parameters for analysis of Bos short reads are not provided. The version and parameters for pbsv analysis of Bos long reads are not provided. The software for determining the overlap between TE detectors and simulated data is not described. The software and specific criteria for determining the concordance between short-read and long-read predictions for Bos datasets are not given.
- Are the methods and statistical analyses appropriate and well described?[ ] Yes, [x] No (please explain), [ ] I don’t know
See above.
Results
- In the case of negative results, is there a statistical power analysis (or an adequate Bayesian analysis or equivalence testing)? [ ] Yes, [ ] No (please explain), [x] I don’t know
Are the results described and interpreted correctly? [ ] Yes, [x] No (please explain), [ ] I don’t know
It would be helpful for all figures to present Drosophila and Arabidopsis data in the same order. (i.e., make fig 3 and 6 like figs 4, 5, 7 & 8.); It is unclear in Fig 9 which format the TE library is in (LTR and internal combined or separate?). The data in Figure 10 do not support the claim that class I ERVs are “better recognized” than class II ERVs. The lack of access to data and detailed description of methods makes it difficult to evaluate claims about short-read TE detector performance on Bos real data. The authors do not seem to be aware that some TE detection systems do not attempt to make reference TE predictions (i.e., TEbreak and Retroseq).
Discussion
- Have the authors appropriately emphasized the strengths and limitations of their study/theory/methods/argument? [ ] Yes, [x] No (please explain), [ ] I don’t know
No comparison to prior TE simulation frameworks is provided. Additionally, unrealistic aspects of the replicaTE system are not discussed (i.e., generation of wholly artificial genomes; generation of TE copy size from an exponential distribution – LTR and TIR elements typically insert with a characteristic size; use of longest TE to represent ancestor – this should be a consensus; access to reference genomes with comprehensive TE annotation as input to simulation).
- Are the conclusions adequately supported by the results (without overstating the implications of the findings)? [x] Yes, [ ] No (please explain), [ ] I don’t know
General comments:
Verneret et al present a new system for the simulation of TE sequences in eukaryotic genomes, together with a performance analysis of several short-read TE detection systems in Drosophila, Arabidopsis and Bos on simulated data, followed by an analysis of real Bos data based on simulation results. The simulation system generates synthetic genomes with characteristics of real genomes that are present in GenBank. There is no attempt to compare this system to previous simulation systems that have been used to evaluate short-read TE detection software (i.e., simulaTE by Kofler 2018 and the single TE insertion framework by Nelson et al 2017/Chen et al 2023). A full treatment of prior work and the strengths and weaknesses (e.g., limited to reference genomes in Genbank, limited number of TE insertions, generation of completely artificial genomes) of replicaTE vis-à-vis prior studies would benefit the reader greatly. The analysis of TE detection system perofmance is a valuable addition to the field and underscores many themes that have been reported in prior work by Rishiwara et al 2016, Nelson et al 2017, Vendrill-Mir et al 2019 and Chen et al 2023. The manuscript (and readers) would benefit from more effort to synthesize similarities and differences between the current and prior work (e.g., Chen et al. 2023 also report that 50X coverage is recommended for the optimal detection of non-reference insertions). Limitations of the current evaluation study should also me more thoroughly discussed (i.e., the authors only analyze a single simulation replicate for each species, and thus quantitative differences among TE detection methods may reflect results for only this replicate). Lastly, the analysis of Bos data is lacking a direct investigation of sequence characteristics that affect short-read TE detection performance (as is shown for Drosophila and Arabidopsis), as well as key information about access to empirical datasets and methodology to reproduce main findings.
https://doi.org/10.24072/pci.genomics.100418.rev11










