
To avoid biases and to be FAIR, we need to CARE and share biodiversity metadata
 based on reviews by Julian Osuji and 1 anonymous reviewer
based on reviews by Julian Osuji and 1 anonymous reviewer
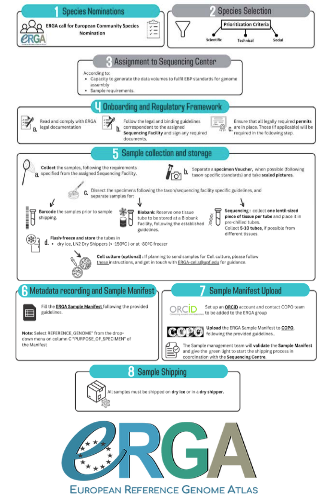
Contextualising samples: Supporting reference genomes of European biodiversity through sample and associated metadata collection
Abstract
Recommendation: posted 28 May 2024, validated 01 July 2024
Sabot, F. (2024) To avoid biases and to be FAIR, we need to CARE and share biodiversity metadata. Peer Community in Genomics, 100255. 10.24072/pci.genomics.100255
Recommendation
Böhne et al. (2024) do not present a classical scientific paper per se but a report on how the European Reference Genome Atlas (ERGA) aims to deal with sampling and sample information, i.e. metadata.
As the goal of ERGA is to provide an almost fully representative set of reference genomes representative of European biodiversity to serve many research areas in biology, they have to be really exhaustive. In this regard, in addition to providing sample metadata recording guidelines, they also discuss the biases existing in sampling and sequencing projects.
The first task for such a project is to be sure that the data they generate will be usable and available in the future (“[in] perpetuity", Böhne et al. 2024). The authors deployed a very efficient pipeline for conserving information on sampling: location, physical information, copies of tissues and of DNA, shipping, legal/ethical aspects regarding the Nagoya Protocol, etc., alongside a best-practice manual. This effort is linked to practical guides for the DNA extraction of specific taxa. More generally, these details enable “Findable, Accessible, Interoperable, and Reusable” (FAIR) principles (Wilkinson et al. 2016) to be followed.
An important aspect of this paper, in addition to practical points, is the reflection upon the different biases inherent to the choice of sequenced samples. Acknowledging their own biases with regards to DNA extraction protocol efficiency, small genome size choice, as well as the availability of material (Nagoya Protocol aspects) and material transfer efficiency, the authors recommend in the future to not survey biodiversity by selecting one’s favorite samples or species, but also considering "orphan" taxa. Some of these "orphan" taxonomic groups belong to non-arthropod invertebrates but internal disparities are also prominent within other taxa. Finally, the implementation of the "Collective benefit, Authority to control, Responsibility, and Ethics" (CARE) principles (Carroll et al. 2021) will allow Indigenous rights to be considered when prioritizing samples, and to enable their "knowledge systems to permeate throughout the process of reference genome production and beyond" (Böhne et al. 2024).
Last, but not least, as ERGA, including its Sampling and Sample Processing committee, is a large collective effort, it is very refreshing to read a paper starting with the acknowledgements and the roles of each member.
References
Böhne A, Fernández R, Leonard JA, McCartney AM, McTaggart S, Melo-Ferreira J, Monteiro R, Oomen RA, Pettersson OV, Struck TH (2024) Contextualising samples: Supporting reference genomes of European biodiversity through sample and associated metadata collection. bioRxiv, ver. 3 peer-reviewed and recommended by Peer Community in Genomics. https://doi.org/10.1101/2023.06.28.546652
Carroll SR, Herczog E, Hudson M, Russell K, Stall S (2021) Operationalizing the CARE and FAIR Principles for Indigenous data futures. Scientific Data, 8, 108. https://doi.org/10.1038/s41597-021-00892-0
Wilkinson MD, Dumontier M, Aalbersberg IjJ, Appleton G, Axton M, Baak A, Blomberg N, Boiten J-W, da Silva Santos LB, Bourne PE, Bouwman J, Brookes AJ, Clark T, Crosas M, Dillo I, Dumon O, Edmunds S, Evelo CT, Finkers R, Gonzalez-Beltran A, Gray AJG, Groth P, Goble C, Grethe JS, Heringa J, ’t Hoen PAC, Hooft R, Kuhn T, Kok R, Kok J, Lusher SJ, Martone ME, Mons A, Packer AL, Persson B, Rocca-Serra P, Roos M, van Schaik R, Sansone S-A, Schultes E, Sengstag T, Slater T, Strawn G, Swertz MA, Thompson M, van der Lei J, van Mulligen E, Velterop J, Waagmeester A, Wittenburg P, Wolstencroft K, Zhao J, Mons B (2016) The FAIR Guiding Principles for scientific data management and stewardship. Scientific Data, 3, 160018. https://doi.org/10.1038/sdata.2016.18
The recommender in charge of the evaluation of the article and the reviewers declared that they have no conflict of interest (as defined in the code of conduct of PCI) with the authors or with the content of the article. The authors declared that they comply with the PCI rule of having no financial conflicts of interest in relation to the content of the article.
All funding credits are listed in the acknowledgements section of the manuscript. The entire list could not be pasted here due to character limitations.
Reviewed by anonymous reviewer 1, 15 Apr 2024
I am happy with the edits made by the authors. Many thanks for the work and the contribution made to the community.
Best wishes
Evaluation round #1
DOI or URL of the preprint: https://doi.org/10.1101/2023.06.28.546652
Version of the preprint: 1
Author's Reply, 14 Feb 2024
Decision by Francois Sabot , posted 27 Oct 2023, validated 27 Oct 2023
, posted 27 Oct 2023, validated 27 Oct 2023
Dear Dr Böhne,
Your paper has been (finally, I am sorry for the delay in finding suitable reviewers), reviewed twice independantly, and I read it myself thouroughly.
It is a very interesting and fundamental "white paper" on this excellent project that is ERGA. However, to have even a greater impact, and particularly in regard of non-core ERGA members, I agreed with the reviewers that some parts need a better explanation (outside of some minor syntax proposals).
Indeed, one may be afraid of the big machine that is ERGA, as previous large initiatives provided data but with "no goals": insisting on the possible usage of these data for the whole scientific community as well as public would be a great improvment of your manuscript.
If you accept to correct this in this regard, I would be pleased to accept the manuscript.
Sincerely yours
Francois Sabot
Reviewed by anonymous reviewer 1, 10 Oct 2023
Reviewed by Julian Osuji, 05 Sep 2023
Review Report
Title:
The title clearly reflects the content of the article. However, I suggest replacement of “for” in the title with “of”
Abstract
The abstract is concise and captures the major points in the article.
55 SSP serves as the sample provider’s entry point… I suggest providers’ reason is that SSP ought to receive several samples; not one sample
I. The Sampling and Sample Processing committee of ERGA
Introduction clearly demonstrates the motivation for the study.
The introduction builds on relevant recent and past reference research.
76-77 Delete the phrase “one of which is the Sampling and Sample Processing committee (SSP)” It seems to appear slightly early. It can come at the beginning sentence of the next paragraph as follows:
88 The Sampling and Sample Processing committee (SSP) is a working group of volunteer expert ERGA members tasked with developing guidelines 83 to support sampling and sample processing.
Materials and Methods
This section contains sufficient information that can be replicated in similar researches.
Results
Data presented in the article are correct and unambiguously presented.
174 ….Widening countries with 44% and 50% of… I suggest … Widening countries with 44 and 50 % of and
175 However, only 36% or 42% of the… However, only 36 or 42 % of the…
The tables and figures (charts) are clear and self-explanatory. However, the texts in Figure 3 could be made more legible for easier reading.
IV. Sample provision: connecting genome teams with 322 sequencing centres
324 arising from three main categories: biological, logistic, and legal issues. I rather think it should be:
324 arising from four main categories: biological, logistic, administrative/policy and legal issues.
364 Future taxon-specific best-practice guidelines
The approach of having different sampling procedures for different taxa is very commendable as it would eliminates complications arising from structural and functional variations between the taxa.
490 References
The listed references are appropriate
General Comment
The article captured very important details associated with an active reference genome community of practice and vividly explained the challenges faced by such a consortium.
Download the review









