
FISTON-LAVIER Anna-Sophie
- Institut des Sciences de l'Evolution de Montpellier (ISEM), Université de Montpellier, Montpellier, France
- Arthropods, Bioinformatics, Evolutionary genomics, Population genomics, Viruses and transposable elements
- recommender, manager, administrator
Recommendations: 2
Reviews: 0
Recommendations: 2

Scalable, accessible and reproducible reference genome assembly and evaluation in Galaxy
A new Galaxy workflow to generate and evaluate reference genome assemblies
Recommended by Alba Marino, Anna-Sophie Fiston-Lavier and Capucine MayoudAlba Marino (1), Capucine Mayoud (1), Anna-Sophie Fiston-Lavier (1,2)
(1) ISEM, Univ Montpellier, CNRS, IRD, Montpellier, France
(2) Institut Universitaire de France
Biodiversity is the bedrock of many ecosystem services fundamental to human society. Acquiring genome-level information appears increasingly important for a deeper understanding of biodiversity and to plan conservation actions for endangered species (Lewin et al. 2022). Consortia such as the Vertebrate Genomes Project (VGP; Rhie et al. 2021) and the European Reference Genome Atlas (ERGA; Formenti et al. 2022) have been undertaken to coordinate global efforts toward sequencing of all the existing vertebrate and European eukaryotic species, respectively. Indeed, generating genome-scale data across such a wide taxonomic range presents significant challenges—not least the development and long-term maintenance of computational tools and workflows that ensure both reproducibility and transparency.
Galaxy offers a user-friendly, web-based environment for executing complex pipelines in a reproducible way, as well as servers for data storage (Bray & Maier 2023). In this context, Larivière et al. (2024) present a major enhancement to reference genome assembly with the development of a scalable, accessible, and reproducible pipeline embedded within the Galaxy platform. The framework has been designed to democratize the production of high-quality genomes, in line with initiatives such as the Earth BioGenome Project (Lewin et al. 2022). It integrates six main stages, namely (1) k-mer genome profiling, (2) phased assembly construction, (3) artefactual duplication purging, (4) scaffolding, (5) decontamination, and (6) mitogenome assembly. The pipeline builds on the expertise of VGP (Rhie et al. 2021) and ERGA (Formenti et al. 2022), while incorporating recent advances in high-fidelity long-read sequencing technologies.
A key strength of the pipeline lies in the open availability and its modularity, which enables end-to-end processing from raw reads to curated assemblies while emphasizing reproducibility, transparency, and ease of use (Afgan et al. 2018). Another major advantage is the integration of quality control steps throughout the pipeline. Moreover, the system is designed to accommodate a wide range of input data types and is applicable to a broad spectrum of species (Larivière et al. 2024).
Several public Galaxy instances are available worldwide (e.g. in the USA: https://usegalaxy.org; in Europe: https://usegalaxy.eu; in Australia: https://usegalaxy.org.au). These platforms provide free access to computing resources for running complex workflows and analysing large datasets. Nonetheless, certain steps in genome assembly may require more memory (RAM) or processing power (CPU) than the instances can offer, thus demanding access to high-performance computing (HPC) environments. Although cloud execution is mentioned as a means of processing large amounts of data, the manuscript offers little detail on deployment costs or potential technical barriers.
Beyond technical and financial considerations, the environmental impact of scaling up genome sequencing and assembly also deserves attention. As more projects are launched and reliance on cloud infrastructure increases, the demand for computing, data storage, and long-term archival will increase substantially. Such operations are energy-intensive and contribute significantly to the environmental footprint of computational biology (Lannelongue & Inouye 2023). While Larivière et al. (2024) rightly emphasize accessibility and scalability, the community must also consider sustainability strategies to limit the ecological impact of large-scale genome initiatives.
The authors suggest that the pipeline can be adapted for non-vertebrate species, such as plants or fungi, by adjusting a few parameters (e.g. BUSCO clade selection). However, the pipeline has so far only been validated on vertebrate genomes. Its robustness across taxa with complex genomic features, such as extreme GC content, polyploidy, or high repeat density, will require further benchmarking. Finally, another challenge is keeping the pipeline up to date. The rapid evolution of genome assembly tools (Nurk et al. 2022) contrasts with the often slower update cycles of Galaxy workflows, raising concerns about maintaining best practice standards without active long-term governance. The pipeline would benefit from an additional step to compare the established Galaxy pipeline with new assembly tools better suited to data generating using the latest technologies.
In conclusion, Larivière et al. (2024) offer a vital step forward in making reference-quality genome assembly broadly accessible. It is now in the hands of the community to address the remaining open challenges, such as computational accessibility, broader taxonomic validation, environmental sustainability, and further proofing of the pipeline.
References
Afgan E, Baker D, Batut B, van den Beek M, Bouvier D, Čech M, Chilton J, Clements D, Coraor N, Grüning BA, Guerler A, Hillman-Jackson J, Hiltemann S, Jalili V, Rasche H, Soranzo N, Goecks J, Taylor J, Nekrutenko A, Blankenberg D (2018) The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Research, 46, W537–W544. https://doi.org/10.1093/nar/gky379
Bray S, Maier W. (2023) Automating Galaxy workflows using the command line. Galaxy Training Network. https://training.galaxyproject.org/training-material/topics/galaxy-interface/tutorials/workflow-automation/tutorial.html
Formenti G, Theissinger K, Fernandes C, Bista I, Bombarely A, Bleidorn C, et al. (2022) The era of reference genomes in conservation genomics. Trends in Ecology & Evolution, 37, 197–202. https://doi.org/10.1016/j.tree.2021.11.008
Lannelongue, L, Inouye, M (2023) Carbon footprint estimation for computational research. Nat Rev Methods Primers 3, 9. https://doi.org/10.1038/s43586-023-00202-5
Larivière D, Abueg L, Brajuka N, Gallardo-Alba C, Grüning B, Ko BJ, Ostrovsky A, Palmada-Flores M, Pickett BD, Rabbani K, Antunes A, Balacco JR, Chaisson MJP, Cheng H, Collins J, Couture M, Denisova A, Fedrigo O, Gallo GR, Giani AM, Gooder GM, Horan K, Jain N, Johnson C, Kim H, Lee C, Marques-Bonet T, O’Toole B, Rhie A, Secomandi S, Sozzoni M, Tilley T, Uliano-Silva M, van den Beek M, Williams RW, Waterhouse RM, Phillippy AM, Jarvis ED, Schatz MC, Nekrutenko A, Formenti G (2024) Scalable, accessible and reproducible reference genome assembly and evaluation in Galaxy. Nature Biotechnology, 42, 367–370. https://doi.org/10.1038/s41587-023-02100-3
Lewin HA, Richards S, Lieberman Aiden E, Allende ML, et al. (2022) The Earth BioGenome Project 2020: Starting the clock. Proceedings of the National Academy of Sciences, 119, e2115635118. https://doi.org/10.1073/pnas.2115635118
Nurk S, Koren S, Rhie A, Rautiainen M, Bzikadze AV, et al. (2022) The complete sequence of a human genome. Science, 376, 44–53. https://doi.org/10.1126/science.abj6987
Rhie A, McCarthy SA, Fedrigo O, Damas J, Formenti G, et al. (2021) Towards complete and error-free genome assemblies of all vertebrate species. Nature, 592, 737–746. https://doi.org/10.1038/s41586-021-03451-0
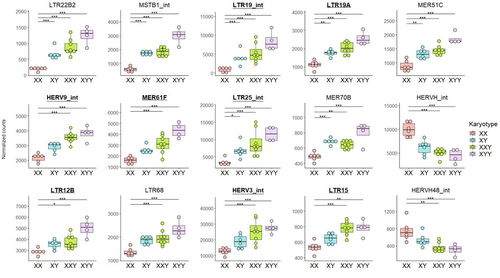
Transposable element expression with variation in sex chromosome number supports a toxic Y effect on human longevity
The number of Y chromosomes is positively associated with transposable element expression in humans, in line with the toxic Y hypothesis
Recommended by Anna-Sophie Fiston-Lavier based on reviews by 3 anonymous reviewersThe study of human longevity has long been a source of fascination for scientists, particularly in relation to the genetic factors that contribute to differences in lifespan between the sexes. One particularly intriguing area of research concerns the Y chromosome and its impact on male longevity. The Y chromosome expresses genes that are essential for male development and reproduction. However, it may also influence various physiological processes and health outcomes. It is therefore of great importance to investigate the impact of the Y chromosome on longevity. This may assist in elucidating the biological mechanisms underlying sex-specific differences in aging and disease susceptibility. As longevity research progresses, the Y chromosome's role presents a promising avenue for elucidating the complex interplay between genetics and aging.
Transposable elements (TEs), often referred to as "jumping genes", are DNA sequences that can move within the genome, potentially causing mutations and genomic instability. In young, healthy cells, various mechanisms, including DNA methylation and histone modifications, suppress TE activity to maintain genomic integrity. However, as individuals age, these regulatory mechanisms may deteriorate, leading to increased TE activity. This dysregulation could contribute to age-related genomic instability, cellular dysfunction, and the onset of diseases such as cancer. Understanding how TE repression changes with age is crucial for uncovering the molecular underpinnings of aging (De Cecco et al. 2013; Van Meter et al. 2014).
The lower recombination rates observed on Y chromosomes result in the accumulation of TE insertions, which in turn leads to an enrichment of TEs and potentially higher TE activity. To ascertain whether the number of Y chromosomes is associated with TE activity in humans, Teoli et al. (2024) studied the TE expression level, as a proxy of the TE activity, in several karyotype compositions (i.e. with differing numbers of Y chromosomes). They used transcriptomic data from blood samples collected in 24 individuals (six females 46,XX, six males 46,XY, eight males 47,XXY and four males 47,XYY). Even though they did not observe a significant correlation between the number of Y chromosomes and TE expression, their results suggest an impact of the presence of the Y chromosome on the overall TE expression. The presence of Y chromosomes also affected the type (family) of TE present/expressed. To ensure that the TE expression level was not biased by the expression of a gene in proximity due to intron retention or pervasive intragenic transcription, the authors also tested whether the TE expression variation observed between the different karyotypes could be explained by gene (i.e. here non-TE gene) expression.
As TE repression mechanisms are known to decrease over time, the authors also tested whether TE repression is weaker in older individuals, which would support a compelling link between genomic stability and aging. They investigated the TE expression differently between males and females, hypothesizing that old males should exhibit a stronger TE activity than old females. Using selected 45 males (47,XY) and 35 females (46,XX) blood samples of various ages (from 20 to 70) from the Genotype-Tissue Expression (GTEx) project, the authors studied the effect of age on TE expression using 10-year range to group the study subjects. Based on these data, they fail to find an overall increase of TE expression in old males compared to old females.
Notwithstanding the small number of samples, the study is well-designed and innovative, and its findings are highly promising. It marks an initial step towards understanding the impact of Y-chromosome ‘toxicity’ on human longevity. Despite the relatively small sample size, which is a consequence of the difficulty of obtaining samples from individuals with sex chromosome aneuploidies, the results are highly intriguing and will be of interest to a broad range of biologists.
References
De Cecco M, Criscione SW, Peckham EJ, Hillenmeyer S, Hamm EA, Manivannan J, Peterson AL, Kreiling JA, Neretti N, Sedivy JM (2013) Genomes of replicatively senescent cells undergo global epigenetic changes leading to gene silencing and activation of transposable elements. Aging Cell, 12, 247–256. https://doi.org/10.1111/acel.12047
Teoli J, Merenciano M, Fablet M, Necsulea A, Siqueira-de-Oliveira D, Brandulas-Cammarata A, Labalme A, Lejeune H, Lemaitre J-F, Gueyffier F, Sanlaville D, Bardel C, Vieira C, Marais GAB, Plotton I (2024) Transposable element expression with variation in sex chromosome number supports a toxic Y effect on human longevity. bioRxiv, ver. 5 peer-reviewed and recommended by Peer Community in Genomics. https://doi.org/10.1101/2023.08.03.550779
Van Meter M, Kashyap M, Rezazadeh S, Geneva AJ, Morello TD, Seluanov A, Gorbunova V (2014) SIRT6 represses LINE1 retrotransposons by ribosylating KAP1 but this repression fails with stress and age. Nature Communications, 5, 5011. https://doi.org/10.1038/ncomms6011