
Comparative genomics in the chestnut blight fungus Cryphonectria parasitica reveals large chromosomal rearrangements and a stable genome organization
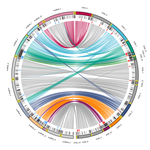
Chromosomal rearrangements with stable repertoires of genes and transposable elements in an invasive forest-pathogenic fungus
Abstract
Recommendation: posted 23 March 2022, validated 23 March 2022
Duplessis, S. (2022) Comparative genomics in the chestnut blight fungus Cryphonectria parasitica reveals large chromosomal rearrangements and a stable genome organization. Peer Community in Genomics, 100013. https://doi.org/10.24072/pci.genomics.100013
Recommendation
About twenty-five years after the sequencing of the first fungal genome and a dozen years after the first plant pathogenic fungi genomes were sequenced, unprecedented international efforts have led to an impressive collection of genomes available for the community of mycologists in international databases (Goffeau et al. 1996, Dean et al. 2005; Spatafora et al. 2017). For instance, to date, the Joint Genome Institute Mycocosm database has collected more than 2,100 fungal genomes over the fungal tree of life (https://mycocosm.jgi.doe.gov). Such resources are paving the way for comparative genomics, population genomics and phylogenomics to address a large panel of questions regarding the biology and the ecology of fungal species. Early on, population genomics applied to pathogenic fungi revealed a great diversity of genome content and organization and a wide variety of variants and rearrangements (Raffaele and Kamoun 2012, Hartmann 2022). Such plasticity raises questions about how to choose a representative genome to serve as an ideal reference to address pertinent biological questions.
Cryphonectria parasitica is a fungal pathogen that is infamous for the devastation of chestnut forests in North America after its accidental introduction more than a century ago (Anagnostakis 1987). Since then, it has been a quarantine species under surveillance in various parts of the world. As for other fungi causing diseases on forest trees, the study of adaptation to its host in the forest ecosystem and of its reproduction and dissemination modes is more complex than for crop-targeting pathogens. A first reference genome was published in 2020 for the chestnut blight fungus C. parasitica strain EP155 in the frame of an international project with the DOE JGI (Crouch et al. 2020). Another genome was then sequenced from the French isolate YVO003, which showed a few differences in the assembly suggesting possible rearrangements (Demené et al. 2019). Here the sequencing of a third isolate ESM015 from the native area of C. parasitica in Japan allows to draw broader comparative analysis and particularly to compare between native and introduced isolates (Demené et al. 2022).
Demené and collaborators report on a new genome sequence using up-to-date long-read sequencing technologies and they provide an improved genome assembly. Comparison with previously published C. parasitica genomes did not reveal dramatic changes in the overall chromosomal landscapes, but large rearrangements could be spotted. Despite these rearrangements, the genome content and organization – i.e. genes and repeats – remain stable, with a limited number of genes gains and losses. As in any fungal plant pathogen genome, the repertoire of candidate effectors predicted among secreted proteins was more particularly scrutinized. Such effector genes have previously been reported in other pathogens in repeat-enriched plastic genomic regions with accelerated evolutionary rates under the pressure of the host immune system (Raffaele and Kamoun 2012). Demené and collaborators established a list of priority candidate effectors in the C. parasitica gene catalog likely involved in the interaction with the host plant which will require more attention in future functional studies. Six major inter-chromosomal translocations were detected and are likely associated with double break strands repairs. The authors speculate on the possible effects that these translocations may have on gene organization and expression regulation leading to dramatic phenotypic changes in relation to introduction and invasion in new continents and the impact regarding sexual reproduction in this fungus (Demené et al. 2022).
I recommend this article not only because it is providing an improved assembly of a reference genome for C. parasitica, but also because it adds diversity in terms of genome references availability, with a third high-quality assembly. Such an effort in the tree pathology community for a pathogen under surveillance is of particular importance for future progress in post-genomic analysis, e.g. in further genomic population studies (Hartmann 2022).
References
Anagnostakis SL (1987) Chestnut Blight: The Classical Problem of an Introduced Pathogen. Mycologia, 79, 23–37. https://doi.org/10.2307/3807741
Crouch JA, Dawe A, Aerts A, Barry K, Churchill ACL, Grimwood J, Hillman BI, Milgroom MG, Pangilinan J, Smith M, Salamov A, Schmutz J, Yadav JS, Grigoriev IV, Nuss DL (2020) Genome Sequence of the Chestnut Blight Fungus Cryphonectria parasitica EP155: A Fundamental Resource for an Archetypical Invasive Plant Pathogen. Phytopathology®, 110, 1180–1188. https://doi.org/10.1094/PHYTO-12-19-0478-A
Dean RA, Talbot NJ, Ebbole DJ, Farman ML, Mitchell TK, Orbach MJ, Thon M, Kulkarni R, Xu J-R, Pan H, Read ND, Lee Y-H, Carbone I, Brown D, Oh YY, Donofrio N, Jeong JS, Soanes DM, Djonovic S, Kolomiets E, Rehmeyer C, Li W, Harding M, Kim S, Lebrun M-H, Bohnert H, Coughlan S, Butler J, Calvo S, Ma L-J, Nicol R, Purcell S, Nusbaum C, Galagan JE, Birren BW (2005) The genome sequence of the rice blast fungus Magnaporthe grisea. Nature, 434, 980–986. https://doi.org/10.1038/nature03449
Demené A., Laurent B., Cros-Arteil S., Boury C. and Dutech C. 2022. Chromosomal rearrangements with stable repertoires of genes and transposable elements in an invasive forest-pathogenic fungus. bioRxiv, 2021.03.09.434572, ver.6 peer-reviewed and recommended by Peer Community in Genomics. https://doi.org/10.1101/2021.03.09.434572
Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, Louis EJ, Mewes HW, Murakami Y, Philippsen P, Tettelin H, Oliver SG (1996) Life with 6000 Genes. Science, 274, 546–567. https://doi.org/10.1126/science.274.5287.546
Hartmann FE (2022) Using structural variants to understand the ecological and evolutionary dynamics of fungal plant pathogens. New Phytologist, 234, 43–49. https://doi.org/10.1111/nph.17907
Raffaele S, Kamoun S (2012) Genome evolution in filamentous plant pathogens: why bigger can be better. Nature Reviews Microbiology, 10, 417–430. https://doi.org/10.1038/nrmicro2790
Spatafora JW, Aime MC, Grigoriev IV, Martin F, Stajich JE, Blackwell M (2017) The Fungal Tree of Life: from Molecular Systematics to Genome-Scale Phylogenies. Microbiology Spectrum, 5, 5.5.03. https://doi.org/10.1128/microbiolspec.FUNK-0053-2016
The recommender in charge of the evaluation of the article and the reviewers declared that they have no conflict of interest (as defined in the code of conduct of PCI) with the authors or with the content of the article. The authors declared that they comply with the PCI rule of having no financial conflicts of interest in relation to the content of the article.
Evaluation round #2
DOI or URL of the preprint: https://doi.org/10.1101/2021.03.09.434572
Version of the preprint: 4
Author's Reply, 16 Feb 2022
Concerning the effector predictions, we corrected all these confusing parts in blue in the text. First, effectors were predicted from all the secreted proteins predicted from SignalP, without considering the likelihood given by SignalP. Second, the 88 CSEP were a subset of these predicted effectors based only on the secreted protein predicted by SignalP with a likelihood greater than 0.5.
We also removed the term « high confidence » associated with this set of effectors and changed the title of FigS5.
hoping these last corrections are correct answers for a PCI genomics recommendation.
Best regards
Cyril Dutech
Decision by Sebastien Duplessis, posted 24 Jan 2022
Dear authors,
After reading in full the latest version of your manuscript, I can see that the remarks from the two reviewers were carefully considered to generate this version, which now reads very well. The revised version of Table 1 with the metrics, now presented for the isolate ESM015, makes the data clearer for the reader.
I only have a final minor concern regarding one of the comments from the second reviewer:
Please explain the following: “EffectorP identified 1,117 models with a putative signal peptide,…pp.”. I don’t think effectorP predicts signal peptides please clarify. This section needs corrections as it is the wrong usage of EffectorP. EffectorP should only be applied on the secretome as mentioned in both original papers. This section says there are 88 high confidence CSEPs. Latter analysis talks about effectors e.g S5 how are all these related?
I had the exact same comment after reading the previous version of the article and unfortunately, this point does not seem to be fully resolved.
EffectorP indeed applies to the secretome, which should be first defined. I am not discussing the best method for secretome prediction, whatever the tools, it remains a prediction from the proteome and any investigator can choose a preferred tool or combination of tools for this. SignalP being one.
Here, I read and understand that SignalP was used on one side and EffectorP on the other one, then the overlap of the two sets is defining the set of high confidence effectors. It is somehow confusing.
One would expect to see i) SignalP applied to the proteome then ii) EffectorP applied onto the predicted secretome, then selection of candidate effectors for further investigation.
Also, in line 1431 the caption for Figure S5 indicates ... Effectors predicted by EffectorP or predicted by SignalP (SP) ... In the latter, they should not be claimed as "effectors" but rather as Predicted Secreted Proteins. This also adds to the confusion as pointed out by the same reviewer.
Finally, on the terms "high confidence effectors". EffectorP remains a prediction tool and high confidence could be gained only by complementary approaches (expression during host-interaction, homology with bona fide effectors, ...). "Selected candidate effectors" would be a more cautious naming. For your information, there is a new version of the EffectorP tool available (if applied, it may confirm the previously unraveled candidates, ... this is not mandatory or a request).
Minor point, CSEP acronym is defined on line 554 and should be used in plain again on line 724.
If the process of selection of your candidates could be clarified, it will be my pleasure to recommend this study in PCIgenomics.
Regards,
Sebastien Duplessis
Evaluation round #1
DOI or URL of the preprint: https://doi.org/10.1101/2021.03.09.434572
Version of the preprint: 3
Author's Reply, 20 Dec 2021
Decision by Sebastien Duplessis, posted 02 Jul 2021
Dear colleagues,
I am happy to send my decision based on the reports from two reviewers who are experts in fungal genomics.
I am sincerely sorry for the too long delay in sending this decision. I was counting on the advices from a third reviewer with strong expertise in the field of forest pathogenomics. Unfortunately, I had to drop my hopes after a long period of silence with no more answers from this colleague. I hope there was no trouble on this reviewer’s side.
After I received the comments from the two reviewers, I went through your manuscript again and I concur with their constructive remarks and I am sure considering these will help strengthen and reshape the manuscript into a great article that I would be happy to recommend to the community!
With my best regards,
Sebastien
Reviewed by anonymous reviewer 1, 18 May 2021
Chromosomal rearrangements are major contributors to genome architecture and evolution but remain overlooked in fungal genomes. This is notably due to the lack of high-quality reference genomes for comparative genomics. In this study, the authors report de novo long-read assembly for the chestnut blight pathogen C. parasitica. The sequenced strain was sampled in the species center of origin (East Asia). The authors produced a high-quality genome assembly almost reaching the chromosome level. If the overall gene and transposable element contents are stable in comparison to the reference genome of a North-American strain, the two strains exhibit extensive chromosomal rearrangements.
This genome report manuscript in itself is solid, the analyses support the results and interpretations.
I have several concerns/comments to address:
1- Why the authors show the assembly metrics of the three strains EP155, YVO003 and ESM015 but do not include YVO003 in the comparative genomic analyses? Including the three strains could already provide some inputs regarding the emergence of asexual reproduction lineages in invasive C. parasitica populations.
2- I do not understand why the authors included failed assemblies in the main Table1 (e.g. Miniasm) and do not provide the final assembly metrics. How is it possible to reach 46Mb assembly and have 0.5% BUSCO, is it an error? A common statement is to consider assembly 'good' is to have at least 95% of completeness with BUSCO. Regarding the HybridSPAdes assembly in Table 1, the minimal (contig/scaffold) length is of 129pb which is really small and I assume prior to curation. Can the authors provide the final (after curation) metrics for the chosen assembly in the main Table1? I would suggest to remove the "working" assemblies from Table 1 (more useful for a supplemental table). That will help the reader to catch the final comparison between the three assemblies of the reference strain EP155, YVO003 and the new strain ESM015.
3- Regarding RIP signatures found in the TEs, do the two strains carry the "RIP-machinery" genes such as DNA methyletransferases (e.g. N. crassa DIM2 gene, or RID gene)? This would strongly support the potential ongoing RIP activity suggested by the RIP-indexes estimations.
4- To improve the manuscript, I think that overall authors could emphasise the interesting findings and questions addressed in the study instead of focusing on 'negative results'. Actually, there are not really "negative results" but the first half of the results of the comparison between the two reference genomes are focusing on: "no two speed genome", 'no variation in effectors or TE repertoires' sound quite negative and kind of mask the main result: stable gene content, extensive rearrangements.
Minor comments
5- Maybe the authors would like to add Badet et al 2020 (https://bmcbiol.biomedcentral.com/articles/10.1186/s12915-020-0744-3 ) as a reference to illustrate how high quality genome assemblies of a variety of strains can unravel structural variation within species.
6- L168-L192: I think this entire paragraph should be removed, I do not think it is needed to define the three generations of sequencing technologies nowadays. I would just mention that high quality assemblies are needed to explore genome architecture, and that's now possible using long-read sequencing. I would just keep the sentence L186-192, which links quite well with the sentence L162-166.
7- L77-78: Is it really necessary to state that some fungi are easy to manipulate in lab conditions? I think it is mostly untrue and that's exactly why genomics are so interesting to perform on such species.
8- L245-246: the types of structural variation were already listed L1.
9- L352: The common usage is to describe the trimming parameters used (quality and length). What is the reference of the prinseq software (the download website is not a reference)?
https://doi.org/10.24072/pci.genomics.100052.rev11Reviewed by Benjamin Schwessinger, 28 May 2021
I am reviewing this manuscript as part of the PCI Genomics initiative. I do apologize being late.
I think the overall conclusion of the manuscript are justified by the presented data that there are some structural variants between the assemblies and some changes in gene content.
I am confused about how candidate effectors were predicted and how CSEPs were analyzed as outlined below.
Major comments:
· I would suggest to tone down the claims on high quality DNA extraction for fungi and nanopore sequencing. The mean read length of 8Kb and N50 15.46kb are good but not outstanding for Nanopore, even for fungi. Also the 260/230 ratio of 1.45 does suggest residual polysaccharides in the DNA prep as this should be closer to 1.8. Plus there are several good fungal DNA extraction protocols available e.g. https://www.protocols.io/workspaces/high-molecular-weight-dna-extraction-from-all-kingdoms/publications and I would suggest the authors to add their protocol to the list.
· This study would benefit with comparison to the recent Stauber et al. work https://elifesciences.org/articles/56279. I presume the current strain would fall into clade CL3 of that paper. This other paper also compared strains against the Crouch et al. 2020 reference but with short reads only.
· Please explain the following: “EffectorP identified 1,117 models with a putative signal peptide,…pp.”. I don’t think effectorP predicts signal pepdites please clarify. This sections needs corrections as it is the wrong usage of EffectorP. EffectorP should only be applied on the secretome as mentioned in both original papers. This sections says there are 88 high confidence CSEPs. Latter analysis talks about effectors e.g S5 how are all these related?
· What is the pre-RIP index? I could not follow the following section:
o “Pre-RIP index was significantly higher than the estimated baseline frequency (1.28 in both the genomes, Figure 3B), suggesting the absence of a RIP signature. By contrast, estimates of the two post-RIP indexes for the four classes of TEs were either significantly higher or lower than the two estimated baseline frequencies (0.67 and 0.82 for TpA/ApT and CpG/GpC respectively, Figure 3B), suggesting a RIP activity for both the two genomes.”
o These two sentences appear to be contradictory to each other.
o I also did not follow the rational for using CpG/GpC for RIP analysis. Please explain.
Minor comments:
· Some of the citations in the literature in the introduction should be updated. E.g. there were several recent pan-genome papers e.g. tomato and soybean who looked at structural variation at the population level.
· The ‘two-speed genome hypothesis’ is only applicable to a subset of oomycete and fungal crop pathogens. There are many crop pathogens that do not show this compartmentalization including rust fungi and others. It would be good to see this reflected int the introduction.
· The DNA extraction protocol and especially the polysaccharide cleanup step is really interesting I would encourage the authors to post a detailed protocol at protocols.io https://www.protocols.io/workspaces/high-molecular-weight-dna-extraction-from-all-kingdoms/publications
· The genome assembly process seems a bit un-orthodox as they lack really good assemblers like Canu, Flye, or Masurca. I am also not convinced the Spades assembly is really the best as the differences in Illumina mapping rates, BUSCOs and Kmers is relatively small and would likely disappear with bootstrapping for these assemblies. Plus the high number of small might be a drawback of this assembly. Have the authors performed some quality control like BlobTools or such to see if these are spurious assemblies of bacterial contaminants? Just a minor comment. Also npScarf is really meant for simpler genomes like bacteria and not eukaryotes.
· Overall nicely curated assembly at the end.
https://doi.org/10.24072/pci.genomics.100052.rev12









