
Exploring genomic determinants of host specialization in Botrytis cinerea
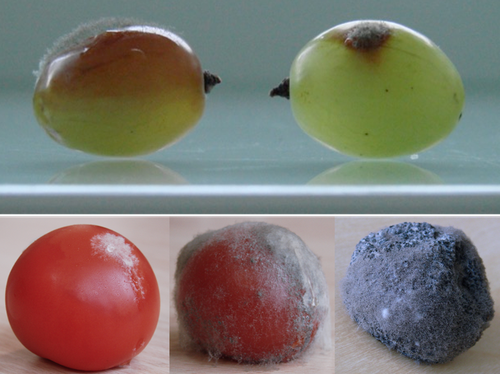
Botrytis cinerea strains infecting grapevine and tomato display contrasted repertoires of accessory chromosomes, transposons and small RNAs
Abstract
Recommendation: posted 14 December 2022, validated 15 December 2022
Duplessis, S. (2022) Exploring genomic determinants of host specialization in Botrytis cinerea. Peer Community in Genomics, . https://doi.org/10.24072/pci.genomics.100023
Recommendation
The genomics era has pushed forward our understanding of fungal biology. Much progress has been made in unraveling new gene functions and pathways, as well as the evolution or adaptation of fungi to their hosts or environments through population studies (Hartmann et al. 2019; Gladieux et al. 2018). Closing gaps more systematically in draft genomes using the most recent long-read technologies now seems the new standard, even with fungal species presenting complex genome structures (e.g. large and highly repetitive dikaryotic genomes; Duan et al. 2022). Understanding the genomic dynamics underlying host specialization in phytopathogenic fungi is of utmost importance as it may open new avenues to combat diseases. A strong host specialization is commonly observed for biotrophic and hemi-biotrophic fungal species or for necrotrophic fungi with a narrow host range, whereas necrotrophic fungi with broad host range are considered generalists (Liang and Rollins, 2018; Newman and Derbyshire, 2020). However, some degrees of specialization towards given hosts have been reported in generalist fungi and the underlying mechanisms remain to be determined.
Botrytis cinerea is a polyphagous necrotrophic phytopathogen with a particularly wide host range and it is notably responsible for grey mould disease on many fruits, such as tomato and grapevine. Because of its importance as a plant pathogen, its relatively small genome size and its taxonomical position, it has been targeted for early genome sequencing and a first reference genome was provided in 2011 (Amselem et al. 2011). Other genomes were subsequently sequenced for other strains, and most importantly a gapless assembled version of the initial reference genome B05.10 was provided to the community (van Kan et al. 2017). This genomic resource has supported advances in various aspects of the biology of B. cinerea such as the production of specialized metabolites, which plays an important role in host-plant colonization, or more recently in the production of small RNAs which interfere with the host immune system, representing a new class of non-proteinaceous virulence effectors (Dalmais et al. 2011; Weiberg et al. 2013).
In the present study, Simon et al. (2022) use PacBio long-read sequencing for Sl3 and Vv3 strains, which represent genetic clusters in B. cinerea populations found on tomato and grapevine. The authors combined these complete and high-quality genome assemblies with the B05.10 reference genome and population sequencing data to perform a comparative genomic analysis of specialization towards the two host plants. Transposable elements generate genomic diversity due to their mobile and repetitive nature and they are of utmost importance in the evolution of fungi as they deeply reshape the genomic landscape (Lorrain et al. 2021). Accessory chromosomes are also known drivers of adaptation in fungi (Möller and Stukenbrock, 2017). Here, the authors identify several genomic features such as the presence of different sets of accessory chromosomes, the presence of differentiated repertoires of transposable elements, as well as related small RNAs in the tomato and grapevine populations, all of which may be involved in host specialization. Whereas core chromosomes are highly syntenic between strains, an accessory chromosome validated by pulse-field electrophoresis is specific of the strains isolated from grapevine. Particularly, they show that two particular retrotransposons are discriminant between the strains and that they allow the production of small RNAs that may act as effectors. The discriminant accessory chromosome of the Vv3 strain harbors one of the unraveled retrotransposons as well as new genes of yet unidentified function.
I recommend this article because it perfectly illustrates how efforts put into generating reference genomic sequences of higher quality can lead to new discoveries and allow to build strong hypotheses about biology and evolution in fungi. Also, the study combines an up-to-date genomics approach with a classical methodology such as pulse-field electrophoresis to validate the presence of accessory chromosomes. A major input of this investigation of the genomic determinants of B. cinerea is that it provides solid hints for further analysis of host-specialization at the population level in a broad-scale phytopathogenic fungus.
References
Amselem J, Cuomo CA, Kan JAL van, Viaud M, Benito EP, Couloux A, Coutinho PM, Vries RP de, Dyer PS, Fillinger S, Fournier E, Gout L, Hahn M, Kohn L, Lapalu N, Plummer KM, Pradier J-M, Quévillon E, Sharon A, Simon A, Have A ten, Tudzynski B, Tudzynski P, Wincker P, Andrew M, Anthouard V, Beever RE, Beffa R, Benoit I, Bouzid O, Brault B, Chen Z, Choquer M, Collémare J, Cotton P, Danchin EG, Silva CD, Gautier A, Giraud C, Giraud T, Gonzalez C, Grossetete S, Güldener U, Henrissat B, Howlett BJ, Kodira C, Kretschmer M, Lappartient A, Leroch M, Levis C, Mauceli E, Neuvéglise C, Oeser B, Pearson M, Poulain J, Poussereau N, Quesneville H, Rascle C, Schumacher J, Ségurens B, Sexton A, Silva E, Sirven C, Soanes DM, Talbot NJ, Templeton M, Yandava C, Yarden O, Zeng Q, Rollins JA, Lebrun M-H, Dickman M (2011) Genomic Analysis of the Necrotrophic Fungal Pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLOS Genetics, 7, e1002230. https://doi.org/10.1371/journal.pgen.1002230
Dalmais B, Schumacher J, Moraga J, Le Pêcheur P, Tudzynski B, Collado IG, Viaud M (2011) The Botrytis cinerea phytotoxin botcinic acid requires two polyketide synthases for production and has a redundant role in virulence with botrydial. Molecular Plant Pathology, 12, 564–579. https://doi.org/10.1111/j.1364-3703.2010.00692.x
Duan H, Jones AW, Hewitt T, Mackenzie A, Hu Y, Sharp A, Lewis D, Mago R, Upadhyaya NM, Rathjen JP, Stone EA, Schwessinger B, Figueroa M, Dodds PN, Periyannan S, Sperschneider J (2022) Physical separation of haplotypes in dikaryons allows benchmarking of phasing accuracy in Nanopore and HiFi assemblies with Hi-C data. Genome Biology, 23, 84. https://doi.org/10.1186/s13059-022-02658-2
Gladieux P, Condon B, Ravel S, Soanes D, Maciel JLN, Nhani A, Chen L, Terauchi R, Lebrun M-H, Tharreau D, Mitchell T, Pedley KF, Valent B, Talbot NJ, Farman M, Fournier E (2018) Gene Flow between Divergent Cereal- and Grass-Specific Lineages of the Rice Blast Fungus Magnaporthe oryzae. mBio, 9, e01219-17. https://doi.org/10.1128/mBio.01219-17
Hartmann FE, Rodríguez de la Vega RC, Carpentier F, Gladieux P, Cornille A, Hood ME, Giraud T (2019) Understanding Adaptation, Coevolution, Host Specialization, and Mating System in Castrating Anther-Smut Fungi by Combining Population and Comparative Genomics. Annual Review of Phytopathology, 57, 431–457. https://doi.org/10.1146/annurev-phyto-082718-095947
Liang X, Rollins JA (2018) Mechanisms of Broad Host Range Necrotrophic Pathogenesis in Sclerotinia sclerotiorum. Phytopathology®, 108, 1128–1140. https://doi.org/10.1094/PHYTO-06-18-0197-RVW
Lorrain C, Oggenfuss U, Croll D, Duplessis S, Stukenbrock E (2021) Transposable Elements in Fungi: Coevolution With the Host Genome Shapes, Genome Architecture, Plasticity and Adaptation. In: Encyclopedia of Mycology (eds Zaragoza Ó, Casadevall A), pp. 142–155. Elsevier, Oxford. https://doi.org/10.1016/B978-0-12-819990-9.00042-1
Möller M, Stukenbrock EH (2017) Evolution and genome architecture in fungal plant pathogens. Nature Reviews Microbiology, 15, 756–771. https://doi.org/10.1038/nrmicro.2017.76
Newman TE, Derbyshire MC (2020) The Evolutionary and Molecular Features of Broad Host-Range Necrotrophy in Plant Pathogenic Fungi. Frontiers in Plant Science, 11. https://doi.org/10.3389/fpls.2020.591733
Simon A, Mercier A, Gladieux P, Poinssot B, Walker A-S, Viaud M (2022) Botrytis cinerea strains infecting grapevine and tomato display contrasted repertoires of accessory chromosomes, transposons and small RNAs. bioRxiv, 2022.03.07.483234, ver. 4 peer-reviewed and recommended by Peer Community in Genomics. https://doi.org/10.1101/2022.03.07.483234
Van Kan JAL, Stassen JHM, Mosbach A, Van Der Lee TAJ, Faino L, Farmer AD, Papasotiriou DG, Zhou S, Seidl MF, Cottam E, Edel D, Hahn M, Schwartz DC, Dietrich RA, Widdison S, Scalliet G (2017) A gapless genome sequence of the fungus Botrytis cinerea. Molecular Plant Pathology, 18, 75–89. https://doi.org/10.1111/mpp.12384
Weiberg A, Wang M, Lin F-M, Zhao H, Zhang Z, Kaloshian I, Huang H-D, Jin H (2013) Fungal Small RNAs Suppress Plant Immunity by Hijacking Host RNA Interference Pathways. Science, 342, 118–123. https://doi.org/10.1126/science.1239705
The recommender in charge of the evaluation of the article and the reviewers declared that they have no conflict of interest (as defined in the code of conduct of PCI) with the authors or with the content of the article. The authors declared that they comply with the PCI rule of having no financial conflicts of interest in relation to the content of the article.
Reviewed by Cecile Lorrain, 23 Sep 2022
The authors made a great effort to perform complementary experiments to support the presence of new accessory chromosomes in B. cinerea.
After revisions, all comments and suggestions of both reviewers were addressed. I consider this manuscript to be suitable for publication with PCI recommendation. I have no further comments.
https://doi.org/10.24072/pci.genomics.100023.rev21
Reviewed by Thorsten Langner, 23 Sep 2022
The authors have addressed all my previous comments and I have no additional comments.
I also want to thank the authors for including the CHEF gel analysis (suggested by reviewer 1) in their revision, which is a nice confirmation and adds value to the genomics analysis.
https://doi.org/10.24072/pci.genomics.100023.rev22Evaluation round #1
DOI or URL of the preprint: https://doi.org/10.1101/2022.03.07.483234
Version of the preprint: 2
Author's Reply, 13 Sep 2022
Decision by Sebastien Duplessis, posted 24 May 2022
Dear authors,
You will find here two complementary reviews of your manuscript. I concur with both reviewers to acknowledge the quality of the work, analyses and interpretations done; also, the text reads very well and only a few edits are suggested.
As you will see, both reviewers provide suggestions to support further some of your analyses or claims (or eventually to revisit and/or tone down some of said claims).
During my first read, I had questions relating to reviewer 1's comments about the use of the GC% signature. Then after another round, I found the comment of reviewer 2 also very pertinent regarding how the age of TE may affect the quality of mapping, and indeed more precisions about the QC you used would be welcome. Finally, both reviewers propose means for managing their comments which will be helpful to reach a manuscript that could be recommended. I am interested in your answers to the first main comment from reviewer 2. As for the first comment from reviewer 1, I do not know how easy this can be routinely handled in the authors' laboratory?
I am looking forward to reading a revised version of this manuscript and your answers to the constructive comments from these reviewers.
With my best regards.
Sebastien
Reviewed by Cecile Lorrain, 13 Apr 2022
Accessory chromosomes in microbes are well-known drivers of evolution and rapid adaptation in many species. Botrytis cinerea strains infecting grapevine and tomato display contrasted repertoires of accessory chromosomes, transposons and small RNAs. The authors have identified new accessory chromosomes in this species, highlighting the importance of high quality sequencing to unravel fungal evolution and biology.
The manuscript is clearly written and the results are well-supported by the produced genomic data and in depth analyses.
I have two main recommendations to improve the manuscript:
1- To further support the presence of accessory chromosomes in the newly sequenced strains, the authors could perform a pulsed-field gel electrophoresis. This potentially would compensate for the scarcity of telomeric repeats in some chromosomes.
2- RIP and GC content: even though RIPped sequenced have low GC content, GC content alone is not the best signature for RIP, since many species exhibit TEs with low %GC but do not have RIP. It can reflect the tendency of TEs to accumulate mutations over time. I suggest the authors, if they want to emphasize the presence of RIP signatures on TEs, to run The RIPper (https://theripper.hawk.rocks/#/home). This user-friendly tool search for RIP indexes in whole-genome or TEs sequences. Also, authors could search for the presence of DNMTs (especially dim2 and RID) genes which are know to be involved in RIP in fungi.
Minor:
- In line 31-33, the authors state that generalists could actually be represented by different co-existing populations specialised in infecting different host species. It would be nice to have the demonstration that it is the case for B. cinerea, just right after the scenario statement. The demonstration comes only from lines 49-61. I would move this part line 36 and finish the paragraph with the different gene catalogues.
-L421-423: remove sentence "Both inactive and active TEs can additionally generate genomic rearrangements by homologous recombination as discussed below."
Reviewed by Thorsten Langner, 07 Apr 2022
The study “Botrytis cinerea strains infecting grapevine and tomato display contrasted repertoires of accessory chromosomes, transposons and small RNAs” describes the genomic features of three genetically diverse, host-adapted lineages of B. cinerea, including a novel accessory chromosome in the grapevine infecting G1 lineage. The authors further show that certain classes of transposable elements are enriched or specific to this lineage and suggest that transposon-derived small RNAs might act as lineage specific effectors. While transposon-derived siRNAs and their potential role in host specificity of B. cinerea have already been described in the literature, the novelty in this study is to extend the analysis to larger numbers of isolates which allows conclusions on the population scale and representative chromosome level assemblies, which allowed to associate certain TE classes to specific chromosomal locations including a novel accessory chromosome. Overall, this study is well done, and most conclusions are supported by the data that is presented.
I only have a few comments that I suggest should be addressed:
Page 15, line 304:
Here, the authors state that predominantly TEs with high GC content produce small RNAs and this appears again in the discussion, but the data is not shown. In my opinion this is an interesting finding which should be supported by data. Otherwise, it is hard to tell how clear the correlation between GC content and small RNA expression is, especially given that the majority of TEs appears to be in the >40%GC category (Figure S11). In addition, the spatial distribution of siRNA-producing TEs in the chromosome level assemblies could add additional value. I suggest to either show this type of analysis or to remove statements and conclusions that are not supported by the results.
I also wonder if the age of TEs and extent of RIP influences the mapping quality of TE derived small RNAs to the consensus and the filtering/masking. This could potentially have an important impact on the overall conclusions of the small RNA producing TEs. Could the authors please comment which quality controls they have done? In the methods section it says, “Quality controls have been done, including PCA…”, however it is hard to tell if any of these controls address the impact of mapping/filtering (e.g. how many reads that map to TEs were discarded during the filtering process? Among those reads, is there a bias that could influence conclusions about GC richness/small RNA expression?).
Page 17, line 348 onwards:
The PCR results and the mapping results are somewhat inconsistent for some isolates. Mapping shows presence of gypsy6 across all tested G1 isolates (Vv1,Vv3,Vv5,Vv14) but PCR fails to detect gypsy6 in these strains. I think the PCR results should thus be interpreted with caution, as presence/absence analysis by PCR can sometimes be problematic. This discrepancy should at least be discussed. Additionally, there are only few details regarding the PCR assay in the paper. It would help if the authors could provide the template sequences used to design the primers and variation of TEs (e.g. alignment of identified TEs similar to what they have done in Figure S8 for the Boty TE, with focus on the primer binding sites). Could variation of TE sequences contribute to absence of PCR products? This could explain the differences between mapping and PCR detection.
Minor comments:
Figure S3: Colors of lines and dots should be explained in the figure legend. Similarly, solid and dashed lines should be explained.
Page 11, line 202: The abbreviation “IPR domains” should be spelled out.
Page 12, line 213: “Fifteen consensus were previously identified” should probably read ”…consensus sequences…”?
Page 19, lines 388/390: These two sentences read a bit contradictory. Maybe it would be worth rephrasing this to point out the lineage specificity of the acc. Chr BCIN19 as a clear difference to 17/18 for which lineage specificity was not supported.
Page 15, lines 291 and following“…producing the higher amount of small RNAs…” should be changed to “…highest amount..”
https://doi.org/10.24072/pci.genomics.100159.rev12









