

 based on reviews by Vittorio Boscaro and 2 anonymous reviewers
based on reviews by Vittorio Boscaro and 2 anonymous reviewers
Several variations of the “universal” genetic code are known. Among the most striking are those where a codon can either encode for an amino acid or a stop signal depending on the context. Such ambiguous codes are known to have evolved in eukaryotes multiple times independently, particularly in ciliates – eight different codes have so far been discovered (1). We generally view such genetic codes are rare ‘variants’ of the standard code restricted to single species or strains, but this might as well reflect a lack of study of closely related species. In this study, Seah and co-authors (2) explore the possibility of codon reassignment in karyorelict ciliates closely related to Parduczia sp., which has been shown to contain an ambiguous genetic code (1). Here, single-cell transcriptomics are used, along with similar available data, to explore the possibility of codon reassignment across the diversity of Karyorelictea (four out of the six recognized families). Codon reassignments were inferred from their frequencies within conserved Pfam (3) protein domains, whereas stop codons were inferred from full-length transcripts with intact 3’-UTRs.
Results show the reassignment of UAA and UAG stop codons to code for glutamine (Q) and the reassignment of the UGA stop codon into tryptophan (W). This occurs only within the coding sequences, whereas the end of transcription is marked by UGA as the main stop codon, and to a lesser extent by UAA. In agreement with a previous model proposed that explains the functioning of ambiguous codes (1,4), the authors observe a depletion of in-frame UGAs before the UGA codon that indicates the stop, thus avoiding premature termination of transcription. The inferred codon reassignments occur in all studied karyorelicts, including the previously studied Parduczia sp. Despite the overall clear picture, some questions remain. Data for two out of six main karyorelict lineages are so far absent and the available data for Cryptopharyngidae was inconclusive; the phylogenetic affinities of Cryptopharyngidae have also been questioned (5). This indicates the need for further study of this interesting group of organisms. As nicely discussed by the authors, experimental evidence could further strengthen the conclusions of this paper, including ribosome profiling, mass spectrometry – as done for Condylostoma (1) – or even direct genetic manipulation.
The uniformity of the ambiguous genetic code across karyorelicts might at first seem dull, but when viewed in a phylogenetic context character distribution strongly suggest that this genetic code has an ancient origin in the karyorelict ancestor ~455 Ma in the Proterozoic (6). This ambiguous code is also not a rarity of some obscure species, but it is shared by ciliates that are very diverse and ecologically important. The origin of the karyorelict code is also intriguing. Adaptive arguments suggest that it could confer robustness to mutations causing premature stop codons. However, we lack evidence for ambiguous codes being linked to specific habitats of lifestyles that could account for it. Instead, the authors favor the neutral view of an ancient “frozen accident”, fixed stochastically simply because it did not pose a significant selective disadvantage. Once a stop codon is reassigned to an amino acid, it is increasingly difficult to revert this without the deleterious effect of prematurely terminating translation. At the end, the origin of the genetic code itself is thought to be a frozen accident too (7).
References
1. Swart EC, Serra V, Petroni G, Nowacki M. Genetic codes with no dedicated stop codon: Context-dependent translation termination. Cell 2016;166: 691–702. https://doi.org/10.1016/j.cell.2016.06.020
2. Seah BKB, Singh A, Swart EC (2022) Karyorelict ciliates use an ambiguous genetic code with context-dependent stop/sense codons. bioRxiv, 2022.04.12.488043. ver. 4 peer-reviewed and recommended by Peer Community in Genomics. https://doi.org/10.1101/2022.04.12.488043
3. Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar GA, Sonnhammer ELL, Tosatto SCE, Paladin L, Raj S, Richardson LJ, Finn RD, Bateman A. Pfam: The protein families database in 2021, Nuc Acids Res 2020;49: D412-D419. https://doi.org/10.1093/nar/gkaa913
4. Alkalaeva E, Mikhailova T. Reassigning stop codons via translation termination: How a few eukaryotes broke the dogma. Bioessays. 2017;39. https://doi.org/10.1002/bies.201600213
5. Xu Y, Li J, Song W, Warren A. Phylogeny and establishment of a new ciliate family, Wilbertomorphidae fam. nov. (Ciliophora, Karyorelictea), a highly specialized taxon represented by Wilbertomorpha colpoda gen. nov., spec. nov. J Eukaryot Microbiol. 2013;60: 480–489. https://doi.org/10.1111/jeu.12055
6. Fernandes NM, Schrago CG. A multigene timescale and diversification dynamics of Ciliophora evolution. Mol Phylogenet Evol. 2019;139: 106521. https://doi.org/10.1016/j.ympev.2019.106521
7. Crick FH. The origin of the genetic code. J Mol Biol. 1968;38: 367–379. https://doi.org/10.1016/0022-2836(68)90392-6
DOI or URL of the preprint: https://doi.org/10.1101/2022.04.12.488043
Version of the preprint: 2
 , posted 09 Jun 2022
, posted 09 Jun 2022Dear authors,
Thank you for submitting this interesting piece of work to PCI Genomics. I very much enjoyed reading this study and as you have probably seen, all three Reviewers were very positive about it too. They also highlighted some minor aspects that could be improved. In particular, please check the possible mislabeling in Figure 2 and consider the confirmation of species IDs using molecular data, which is an important aspect and should be easy to do with the new RNAseq data.
From my side, I have two additional minor comments:
Lines 79-81. I am not sure I fully understand this sentence, could you please clarify what is meant by the frequencies of stop codons within conserved protein domains falling “within the range of observed for coding codons in organisms with known genetic codes”?
Figure 4C. Could you please name the X axis?
Provided an appropriate response to our comments, I would be more than happy to recommend this study. If possible, please enclose a point-by-point response to all the comments.
Sincerely,
Iker Irisarri
The paper by Seah, Singh, & Swart reports an already known but intriguing phenomenon in a large number of other ciliate species, which are good representative of two classes of phylogenetic (and hence evolutionary) interest. The manuscript is very straightforward, the scope is a bit narrow but with strongly supported conclusions, and the figures and text are clear. I also think the authors do a good job at explaining context-dependent codon usage to readers outside the field, and at describing the evolutionary framework they are found in. I have a single main comment, and a series of very minor suggestions that the authors might wish to consider.
Also, while the structure of the manuscript is sound and well thought out, there are occasional sentences that are a bit harder on the reader. This is partly because some of the essential concepts in this work have intrinsically confusing names (e.g. actually terminating stop codons vs. coding stop codons). Some examples are given below, but one suggestion that might fix many issues would be to spell out the subject of sentences whenever feasible, minimizing the instanced of “this” “these”, “it” etc.
MAIN COMMENT
Ideally, the authors should provide a bit more data/info on the way they IDed the ciliates. All I could find was “ciliate cells were identified by morphology under a dissection microscope” (lines 288-289). The authors only go as far down as genera with their assignments, so in a pinch this might do, since genera are usually easy to identify in both heterotrichs and karyorelicts. However, there is some room for uncertainty, and results on imprecisely assigned specimens has long-reach consequences, even if it does not impact the conclusions of this paper.
The authors should have access to the 18S rRNA gene sequences of the specimens they isolated. I would suggest to deposit these separately, and explicitly state that they were used to confirm the morphological assignments. At a minimum, the authors should use BLASTN similarities. Preferably, they should build a small phylogenetic tree by adding their sequences to reference heterotrichs and karyorelicts (e.g., taken from the PR2/EukRef database). Please note I do not suggest that the authors bog down their result section with this. A few sentences in the methods and a tree as a Supplementary Figure would suffice.
MINOR SUGGESTIONS
Lines 23, 59, and 216: While Karyorelictea are certainly fascinating, and they are indeed globally distributed and probably under-sampled, it is a bit of a stretch to call them “abundant” – most ciliates are ecologically important for one reason or another, but only rarely in terms of their number and biomass. Maybe highlight a different trait, or provide a citation somewhere in the text about their underappreciated abundance?
Line 42: “alveolate” and “ciliate” refer to different taxonomic ranks, so I would not use them in the same sentence in opposition. I suggest saying “dinoflagellate” for Amoebophrya.
Lines 60-62: It is not essential and there is no obvious causal connection with the genetic code, but maybe the authors could also mention here that karyorelicts also differ from all other ciliates in their macro/micronuclear cycle pattern.
Lines 70-71: please change to “… 25 transcriptome assemblies (15 [of which] previously published) were used to…” for symmetry.
Lines 89-90: an example of a sentence where “these” is a bit ambiguous.
Line 97: The reference to Figure 4D is out of order compared to all other figures.
Line 100: provide a citation for BUSCO and the Alveolata marker set you used.
Lines 147-150: I don’t understand this sentence. I guess that my main issue is understanding how the second part logically follows the first?
Line 181: “and” instead of “while”, maybe?
Lines 212-214: feel free to mention that Wilbertomorphidae are also monotypic and, to my knowledge, only observed once. It is absolutely understandable that the sort of data needed for this paper is missing for this family.
Lines 260-278: no comments here, I just want to give credit to the authors for highlighting how facile adaptationist speculations can be – and clarifying that one would need evidence to claim there is an adaptive value in any trait in the first place. Very weak adaptive explanations have been proposed for other genomic processes in ciliates in the past.
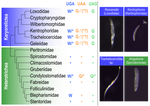
FIGURE 1: “Trachelocercidae” and “sp.” should not be in italics. Also, please move the two Blepharisma entries next to each other.
FIGURE 2: I would suggest not using italics in certain labels, such as “In-frame UGA” or in the Library source box.
FIGURE 3: in here, genera should be italicized. Also, shouldn’t the codons related to each column be shown somehow?
FIGURE 6: the layout of the right side of this figure suggests, at first glance, that Remanella and Kentrophoros are karyorelicts, while Trachelocerca and Anigsteinia are heterotrichs… Also notice that elsewhere in the paper you only mention about having collected Trachelocercidae, not specifically Trachelocerca, which is a trickier claim (see my main comment, but in this case probably not even the 18S would suffice, since genera in Trachelocercidae are probably non-monophyletic).
Seah et al. assessed the genetic codes of two ciliate groups (karyorelicts and heterotrichs) using existing and newly generated genomic/transcriptomic data, and show that karyorelicts use an ambiguous stop/sense codons. This study should be of broad interest to geneticists and protistologists, and will be helpful for genome annotations of ciliates (as well as other eukaryotes). I think the manuscript is well written, the analyses are well-designed and appropriately performed, and all code and raw data are publicly available. I congratulate the authors on a very nice manuscript!
I only have a couple of minor suggestions that the authors might want to consider for improving their manuscript.
L 89-90: "Nonetheless, these were all still...". What do the authors mean by "these"?
L 96-97: It would be interesting to know if the percentage of transcripts with in-frame UGAs is impacted by genome completeness. Do the authors investigate this?
Figure 3: I found this figure to be a difficult to read. First, "codons with frequencies less than 0.02 are highlighted in red". I did not see this at first, and I wonder if this can be made more obvious somehow. Second, I think it would be helpful to have an axis at the bottom with the indicating the codon under consideration.
After generating ten new single-cell RNA-seq libraries, Kwee Boon Seah and colleagues performed an in-depth computational analysis to infer the genetic code of a number of karyorelict and heterotrich ciliates. In continuity with Swart et al 2016, this work expands our knowledge about alternative nuclear genetic codes and provides additional evidence about the existence of a context-dependent ambiguous genetic code in karyorelicts ciliates. While lacking some direct experimental evidence (see below) and mechanistic insights, the genomic analysis is carefully conducted and the results are compelling and convincingly discussed. Overall the paper reads very well and I believe it could be of interest for a broad scientific audience. However, I have some minor comments that should be addressed: