
POOTAKHAM Wirulda
- National Omics Center, National Science and Technology Development Agency, Khlong Luang, Thailand
- Plants
- recommender
Recommendations: 3
Reviews: 0
Recommendations: 3
Natural variation in chalcone isomerase defines a major locus controlling radial stem growth variation among Populus nigra populations
Advancing our understanding of poplar growth using a multi-omics approach
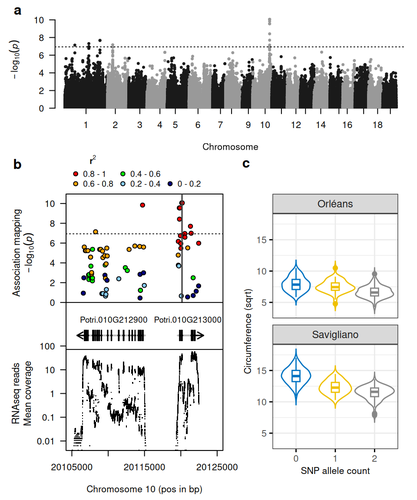
Recommended by Wirulda Pootakham based on reviews by Gancho Slavov and 1 anonymous reviewerPoplar is a promising resource, valued not only for wood production and the development of lignocellulosic biomass, but also for its potential role in carbon sequestration. Recognizing the importance of stem growth for wood production and biomass development, Duruflé et al. (2025) present a comprehensive study on the genetic basis of radial stem growth variation in natural populations of black poplar (Populus nigra). They employed a systems biology approach to identify the quantitative trait loci (QTLs) underlying this trait, integrating genomic, transcriptomic, and phenotypic data from a large collection of poplar genotypes. Their genome-wide association study (GWAS) analysis identified single nucleotide polymorphisms linked to two gene models predicted to encode chalcone isomerase, an enzyme involved in the flavonoid pathway. The authors then used the RNA-seq data to test whether the expression of the candidate genes correlated with the phenotypes, and indeed the level of expression of both genes displayed a correlation to the stem circumference. To support their findings, the authors compared the location of the QTLs detected in this study with previously published QTLs. Interestingly, they found a previously reported QTL co-localizing with the newly identified one. The authors have addressed the concerns raised by reviewers on the GWAS analysis and discussed the complication of this QTL study in the manuscript.
In essence, the authors have combined the power of GWAS and transcriptomics to locate candidate genes and applied population genetics to explore the evolutionary context of the identified gene. This comprehensive approach provides strong evidence for the role of chalcone isomerase in controlling radial stem growth variation in black poplar. The study opens up avenues for further research into the precise mechanisms by which chalcone isomerase and flavonoid metabolism influence stem growth and provides useful information for future poplar breeding programs.
References
Duruflé H, Déjardin A, Jorge V, Pégard M, Pilate G, Rogier O, Sanchez L, Segura V (2025) Natural variation in chalcone isomerase defines a major locus controlling radial stem growth variation among Populus nigra populations. bioRxiv, ver. 3 peer-reviewed and recommended by PCI Genomics. https://doi.org/10.1101/2024.10.21.618920
Phenotypic and transcriptomic analyses reveal major differences between apple and pear scab nonhost resistance
Apples and pears: two closely related species with differences in scab nonhost resistance
Recommended by Wirulda Pootakham based on reviews by 3 anonymous reviewersNonhost resistance is a common form of disease resistance exhibited by plants against microorganisms that are pathogenic to other plant species [1]. Apples and pears are two closely related species belonging to Rosaceae family, both affected by scab disease caused by fungal pathogens in the Venturia genus. These pathogens appear to be highly host-specific. While apples are nonhosts for Venturia pyrina, pears are nonhosts for Venturia inaequalis. To date, the molecular bases of scab nonhost resistance in apple and pear have not been elucidated.
This preprint by Vergne, et al (2022) [2] analyzed nonhost resistance symptoms in apple/V. pyrina and pear/V. inaequalis interactions as well as their transcriptomic responses. Interestingly, the author demonstrated that the nonhost apple/V. pyrina interaction was almost symptomless while hypersensitive reactions were observed for pear/V. inaequalis interaction. The transcriptomic analyses also revealed a number of differentially expressed genes (DEGs) that corresponded to the severity of the interactions, with very few DEGs observed during the apple/V. pyrina interaction and a much higher number of DEGs during the pear/V. inaequalis interaction.
This type of reciprocal host-pathogen interaction study is valuable in gaining new insights into how plants interact with microorganisms that are potential pathogens in related species. A few processes appeared to be involved in the pear resistance against the nonhost pathogen V. inaequalis at the transcriptomic level, such as stomata closure, modification of cell wall and production of secondary metabolites as well as phenylpropanoids. Based on the transcriptomics changes during the nonhost interaction, the author compared the responses to those of host-pathogen interactions and revealed some interesting findings. They proposed a series of cascading effects in pear induced by the presence of V. inaequalis, which I believe helps shed some light on the basic mechanism for nonhost resistance.
I am recommending this study because it provides valuable information that will strengthen our understanding of nonhost resistance in the Rosaceae family and other plant species. The knowledge gained here may be applied to genetically engineer plants for a broader resistance against a number of pathogens in the future.
References
1. Senthil-Kumar M, Mysore KS (2013) Nonhost Resistance Against Bacterial Pathogens: Retrospectives and Prospects. Annual Review of Phytopathology, 51, 407–427. https://doi.org/10.1146/annurev-phyto-082712-102319
2. Vergne E, Chevreau E, Ravon E, Gaillard S, Pelletier S, Bahut M, Perchepied L (2022) Phenotypic and transcriptomic analyses reveal major differences between apple and pear scab nonhost resistance. bioRxiv, 2021.06.01.446506, ver. 4 peer-reviewed and recommended by Peer Community in Genomics. https://doi.org/10.1101/2021.06.01.446506

Gut microbial ecology of Xenopus tadpoles across life stages
A comprehensive look at Xenopus gut microbiota: effects of feed, developmental stages and parental transmission
Recommended by Wirulda Pootakham based on reviews by Vanessa Marcelino and 1 anonymous reviewerIt is well established that the gut microbiota play an important role in the overall health of their hosts (Jandhyala et al. 2015). To date, there are still a limited number of studies on the complex microbial communites inhabiting vertebrate digestive systems, especially the ones that also explored the functional diversity of the microbial community (Bletz et al. 2016).
This preprint by Scalvenzi et al. (2021) reports a comprehensive study on the phylogenetic and metabolic profiles of the Xenopus gut microbiota. The author describes significant changes in the gut microbiome communities at different developmental stages and demonstrates different microbial community composition across organs. In addition, the study also investigates the impact of diet on the Xenopus tadpole gut microbiome communities as well as how the bacterial communities are transmitted from parents to the next generation.
This is one of the first studies that addresses the interactions between gut bacteria and tadpoles during the development. The authors observe the dynamics of gut microbiome communities during tadpole growth and metamorphosis. They also explore host-gut microbial community metabolic interactions and demostrate the capacity of the microbiome to complement the metabolic pathways of the Xenopus genome. Although this study is limited by the use of Xenopus tadpoles in a laboratory, which are probably different from those in nature, I believe it still provides important and valuable information for the research community working on vertebrate’s microbiota and their interaction with the host.
References
Bletz et al. (2016). Amphibian gut microbiota shifts differentially in community structure but converges on habitat-specific predicted functions. Nature Communications, 7(1), 1-12. doi: https://doi.org/10.1038/ncomms13699
Jandhyala, S. M., Talukdar, R., Subramanyam, C., Vuyyuru, H., Sasikala, M., & Reddy, D. N. (2015). Role of the normal gut microbiota. World journal of gastroenterology: WJG, 21(29), 8787. doi: https://dx.doi.org/10.3748%2Fwjg.v21.i29.8787
Scalvenzi, T., Clavereau, I., Bourge, M. & Pollet, N. (2021) Gut microbial ecology of Xenopus tadpoles across life stages. bioRxiv, 2020.05.25.110734, ver. 4 peer-reviewed and recommended by Peer community in Geonmics. https://doi.org/10.1101/2020.05.25.110734