

 based on reviews by Jean-Baptiste Ledoux and 3 anonymous reviewers
based on reviews by Jean-Baptiste Ledoux and 3 anonymous reviewers
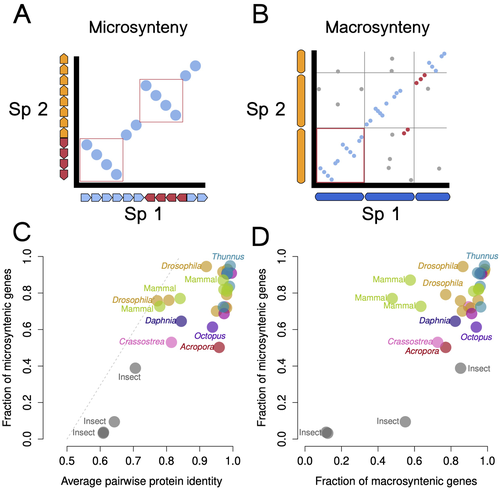
Francis et al. (2025) investigate the relationship between genomic rearrangement, specifically macro- and micro-synteny, and speciation across a broad range of animal phyla. Using chromosome-level genome assemblies, they generated 1:1 ortholog pairs and analyzed synteny conservation using custom bioinformatics pipelines to quantify microsynteny. The study is well written, methodologically sound, and offers valuable insights beyond comparative genomics.
The authors show that while most congeneric species pairs exhibit disruptions in micro-synteny, they retain high levels of protein sequence identity. They also find that macro- and micro-synteny decay with speciation but are often decoupled, indicating no universal genomic trajectory during divergence. Their conclusion, that synteny patterns alone are insufficient to define species boundaries (Steenwyk and King 2024), is well supported by their data.
The discussion effectively situates the work within the broader context of speciation research. It thoughtfully addresses study limitations, such as challenges in synteny block quantification, chromosomal rearrangement rates, and the scarcity of high-quality genome assemblies. The manuscript also outlines clear directions for future research, including the need for more accurate divergence time estimates and expanded taxonomic sampling (Formenti et al. 2022).
References
Formenti G, Theissinger K, Fernandes C, Bista I, Bombarely A, Bleidorn C, et al. (2022) The era of reference genomes in conservation genomics. Trends in Ecology & Evolution, 37, 197–202. https://doi.org/10.1016/j.tree.2021.11.008
Francis WR, Vargas S, Wörheide G (2025) Genomic changes are varied across congeneric species pairs of animals. bioRxiv, ver. 4 peer-reviewed and recommended by PCI Genomics https://doi.org/10.1101/2024.09.05.611358
Steenwyk JL, King N (2024) The promise and pitfalls of synteny in phylogenomics. PLOS Biology, 22, e3002632. https://doi.org/10.1371/journal.pbio.3002632
The authors have performed minor changes. Mainly addressing changing the genus of Crassostrea to Magallana. The title was also modified making it a bit more specific. All of my main concerns were addressed.
https://doi.org/10.24072/pci.genomics.100415.rev31In their rebuttal, the authors explain that, for a variety of reasons, they are unable to make significant changes to their manuscript at this stage. I agree that the two changes (to the title and to oyster nomenclature) that they have made improve the manuscript, but have no further comments at this time because these represent only minimal edits. While I think that addressing more of the comments received would be preferable, I acknowledge the difficulties of this and am happy to recommend the article in its current form. I wish the authors luck with its publication.
https://doi.org/10.24072/pci.genomics.100415.rev32I have no further comments.
Thank you for the opportunity to review this interesting paper.
Best regards,
Jean-Baptiste
DOI or URL of the preprint: https://doi.org/10.1101/2024.09.05.611358
Version of the preprint: 3
 , posted 11 Mar 2025, validated 12 Mar 2025
, posted 11 Mar 2025, validated 12 Mar 2025Dear Dr. Wörheide,
Although all the reviewers found your manuscript valuable and engaging, they still have several comments for improvement. Some reviewers also indicate that some of the suggestions they previously made in the first round of reviews have not been addressed. Therefore, I recommend revising your preprint following the reviewers' suggestions.
Best regards,
Javier del Campo, PhD
I commend the authors for their responses to my comments and for addressing my concerns. I now have only minor comments.
The document with all the supplementary figures greatly facilitated the identification of figures referenced in the manuscript. I thank you for providing it. However, the text within the figures in this document appears slightly blurry. If possible, improving its clarity would enhance readability.
Furthermore, while I understand the authors' perspective that the title is already sufficiently long, I would suggest slightly modifying to make it less generic.
https://doi.org/10.24072/pci.genomics.100415.rev21The authors present an updated version of their manuscript studying genome evolution between congeneric species pairs.
As noted previously, I very much enjoyed reading the manuscript and believe it will be a valuable addition to the field. However, the authors have made minimal efforts to address my comments - which is of course their prerogative - but most of my previous concerns therefore remain and I have little more to add.
https://doi.org/10.24072/pci.genomics.100415.rev22This is the second time I review this paper, entitled “Genomic changes are varied across congeneric species pairs” by Francis and collaborators. In my opinion, the Authors did a good job in considering most of the comments that were made in the previous round.
I still believe that the paper would benefit from the addition of some microevolutionary/ popgen inputs in the introduction and discussion. My comment #4 was indeed not a question but a suggestion in this line. The concept of the “grey zone of speciation” fits with the discussion on divergence time among species etc. but with a population genomics perspective. This may broaden a bit the readership targeted by the manuscript while it supports that this species delimitation is a tricky question at the heart of evolutionary biology.
Anyway, I would like to acknowledge once again the rephrasing and adjustments made by the Authors that significantly clarify the outputs of the study.
Jean-Baptiste
Title and abstract
Does the title clearly reflect the content of the article? Yes
Does the abstract present the main findings of the study? Yes
Introduction
Are the research questions/hypotheses/predictions clearly presented? Yes
Does the introduction build on relevant research in the field? Yes
Materials and methods
Are the methods and analyses sufficiently detailed to allow replication by other researchers? Yes
Are the methods and statistical analyses appropriate and well described? Yes
Results
In the case of negative results, is there a statistical power analysis (or an adequate Bayesian analysis or equivalence testing)? Yes
Are the results described and interpreted correctly? Yes
Discussion
Have the authors appropriately emphasized the strengths and limitations of their study/theory/methods/argument? Yes
Are the conclusions adequately supported by the results (without overstating the implications of the findings)? Yes
https://doi.org/10.24072/pci.genomics.100415.rev24
DOI or URL of the preprint: https://doi.org/10.1101/2024.09.05.611358
Version of the preprint: 1
 , posted 02 Dec 2024, validated 04 Dec 2024
, posted 02 Dec 2024, validated 04 Dec 2024Dear Dr. Wörheide,
Although all the reviewers found your manuscript valuable and interesting, they have provided several comments for improvement. Therefore, I recommend revising your preprint following the reviewers' suggestions
Best regards,
Javier del Campo, PhD
Group Leader, Microbial Ecology and Evolution Laboratory
Chair of the Biodiversity Program
Institut de Biologia Evolutiva (CSIC-UPF)
Passeig Marítim de la Barceloneta 37-49
08003 Barcelona
In this manuscript, entitled “Genomics change are varied across congeneric species pairs”, Francis and collaborators look for a general role of macro- and micro-synteny breaking in speciation across the animal kingdom. To reach this objective, they “examined macro- and micro-synteny in congeneric pairs with chromosome-level genome assemblies representing six animal phyla” adding the newly annotated Acropora hyacinthus genome (l. 70-80 p.2). Following different quality check and data filtering steps, they generated ortholog pairs and quantify micro- and macro-synteny between pair of congeneric species based on home-made bioinformatics pipelines.
Regarding micro-synteny, the Authors found that most species pairs differ in synteny but show relatively high sequence identity; that macro- and microsynteny together tend to decrease with speciation but can be decoupled in many species’ pairs. They discussed the low occurrence of obvious karyotype differences in their dataset. Accounting for contrasted level of divergence between species within pair, the Authors suggest that the pattern of proteins identity values are similar across species pairs. They discussed how the multiple genetic trajectories of genetic change among congeneric species revealed by their study precludes the definition of species delimitations based on patterns of micro- or macro-synteny. Interestingly, they also provided and discussed some of the limitations of their study, including quantification of synteny blocks or estimations of the rates of chromosome rearrangements.
As a conclusion, the Authors stated that “genomic changes inferred from speciation are variable and contextual and thatsynteny should not be used at present as a measure of divergence or speciation ».
My skills in comparative genomics and synteny analyses are relatively limited, so I hope this review to be constructive and relevant.
I found the paper very interesting and clear, even for a non-specialist. The study is quite ambitious and well-conducted. The authors did a great job to involve their work in the broad context of speciation.
Accordingly, the conclusions, i.e. lack of general link between macro-micro-synteny breaking patterns and speciation, are of interest for a broad readership going beyond the comparative geneticists’ community.
Here are some minor comments that hopefully can improve the current version of the MS:
1) The introduction is well written and interesting drawing the broad “speciation” context. Albeit, I would expect a bit more details regarding current knowledge on synteny patterns. For instance, on l. 36 to 55 on page 2, maybe more context should be added to the different examples. Reading (quickly!) Jiang et al. 2019, it is not clear to me how the Authors reached the conclusion that, “extensive syntenic blocks, of tens to hundreds of genes, have been found in octocorals (Jiang et al. 2019) but not medusozoans (Jiang et al. 2019)”.
2) l.5-20 p.3: This paragraph may be improved a bit. It is not clear whether all the insect and mamal pairs involved species from different genus. I suggest to mention this dataset in the Material and Methods. Then regarding the two processes suggested by the Authors, it is not clear whether those are exclusive or not (I guess not) and at stake both in mammals and insects (Ok for insects but do we really have a significant loss of synteny or decrease of protein identity in mammals compared to other species pairs?).
3) Figure 2: Can the Authors justify a bit more the choice of the 4 species pairs represented in the figure? As mentioned, I am not a specialist but I found the identification of red/blue boxes in C and D difficult to understand and poorly justified. It seems to me that many other cells showed similar dot patterns. Maybe, the legend can be improved a bit to facilitate the understanding of the figure.
4) The discussion on the limitations of the study and particularly on the lack of divergence similarity among pairs within group is interesting. There are different papers addressing this topic with a population genetics perspective. They can be of interest to complement the reasoning of the Authors with some “microevolutionary perspectives” (see for instance: Roux C, Fraïsse C, Romiguier J, Anciaux Y, Galtier N, Bierne N (2016) Shedding Light on the Grey Zone of Speciation along a Continuum of Genomic Divergence. PLoS Biol 14(12): e2000234. doi:10.1371/journal.pbio.2000234; see also https://doi.org/10.1016/j.tree.2020.03.002 or Mérot, C., Stenløkk, K.S.R., Venney, C., Laporte, M., Moser, M., Normandeau, E., Árnyasi, M., Kent, M., Rougeux, C., Flynn, J. M., Lien, S., & Bernatchez, L. (2023). Genome assembly, structural variants, and genetic differentiation between lake whitefish young species pairs (Coregonus sp.) with long and short reads. Molecular Ecology, 32, 1458–1477. https://doi.org/10.1111/mec.16468).
5) l.35-37 p.4: “Like the relationship with protein identity, we found that both macrosynteny and microsynteny together tend to decrease with speciation” The meaning of this sentence is not clear. What does speciation mean here? Are the Authors talking about divergence between species within each pair (but see l.14-16 p.5)?
6) I was wondering whether or not it may be of interest to include another layer in the pairwise comparisons by considering, in the different analyses, species pairs which are found in sympatry vs. species pairs found in allopatry. The Authors touched this topic with their comment on Cassostrea species (l. 10-14 p.6).
7) The paragraph entitled “Distribution of protein identify value…” is a bit hard to follow. The link between the expectations (l.78-82 p.4) and the results (l.1-11 p.5) is unclear and could be improved.
Some sentences may also be rephrased. I am probably misunderstanding the reasoning of the Authors but my feeling is that “As mutations accumulate (Figure 3B,C,E,F), very few proteins remain completely identical” is a bit tautologic. More mutation more differences… hmmm? I am probably missing something.
Title and abstract
Does the title clearly reflect the content of the article? Yes,
Does the abstract present the main findings of the study? Yes
Introduction
Are the research questions/hypotheses/predictions clearly presented? Yes
Does the introduction build on relevant research in the field? Yes, but I would like to have a better picture of existing knowledge regarding patterns of synteny.
Materials and methods
Are the methods and analyses sufficiently detailed to allow replication by other researchers? Yes
Are the methods and statistical analyses appropriate and well described? Yes but see comments
Results
In the case of negative results, is there a statistical power analysis (or an adequate Bayesian analysis or equivalence testing)? Not relevant
Are the results described and interpreted correctly? Yes
Discussion
Have the authors appropriately emphasized the strengths and limitations of their study/theory/methods/argument? Yes
Are the conclusions adequately supported by the results (without overstating the implications of the findings)? Yes
This study comprehensively analyzed the micro- and macro-synteny, as well as protein identity, of 1-to-1 orthologs in congeneric species pairs across diverse non-model phyla. By examining these factors, the authors investigated their potential role in speciation. Their findings revealed a correlation between protein identity and microsynteny, but also identified disparities in the conservation of macro- and microsynteny. Moreover, they observed variations in synteny conservation, suggesting a lack of a singular genomic trajectory during speciation.
Although limited by the relatively small dataset of available chromosome-level genome assemblies at the time, this research offers valuable insights into key concepts relevant to future studies on genomic variation and speciation. While the authors were unable to pinpoint a specific genomic feature to delineate species boundaries, they propose that future investigations, addressing limitations such as accurate divergence time estimation independent of protein identity, may uncover discernible patterns. Additionally, this study provides a foundation for future research by bringing together several key concepts and definitions that will be essential for understanding genomic variation in relation to speciation.
I enjoyed reading the manuscript and I only have a few questions and suggestions.
In general, renaming the supplementary figures in the the repositories (github, zenodo) would make following the manuscript easier. Additionally, it seems that they do not appear in the correct order.
In Introduction:
- In page 2, line 52 you mention that you use genomes representing six animal phyla, however I was only able to find five phyla (Mollusca, Chordata, Echinodermata, Arthropoda and Cnidaria). Which would be the sixth?
In Methods - Datasets from NCBI:
- Which is Supplemental Figure 1?
- Adding a supplementary table (expanding Table 1) with all of the genomes used including basic features such as their taxonomic classification, genome ID, assembly and annotation strategies (if available) and genomic features such as genome size, number of chromosomes, % of Ns and BUSCO completeness would be very useful.
In Methods - Generation of ortholog pairs:
- In page 3, line 28: Shouldn’t “For each species” be “For each pair of species”?
- Which is Supplemental Figure 1? Is it the same as the one referenced before in the previous section?
- Why was Crasstostrea used as all vs all and not chose one reference to compare against the others like in Drosophila or Ephinephelus?
- Which is Supplemental Figure 5? Check the order of Supplemental Figures appearances.
- Which test was performed to support the following claim: "The frequency of gaps by species is examined in Supplemental Figure 5 but does not appear to cause any obvious bias."
In Methods - Quantification of microsynteny in genome pairs:
- Please label the Supplementary Figures in the repositories.
In Methods - Quantification of macrosynteny:
- For identifying homologous chromosomes, you only consider a one to one relation between chromosomes in species? This approach would not detect chromosome fusions and fissions. While most of the pairs of species you assessed have the same number of chromosomes this approach could miss fusions and fissions that led to the same number of chromosomes. This would be even more difficult in species with a different number of chromosomes.
In Results - Microsynteny changes more frequently than macrosynteny:
- In Page 4, line 65. How do you distinguish between a fusion and a fission in both Daphnia and Crassostrea without using a third species?
- Page 4, line 67, Crassostea should be Crassostrea.
In Results - Distribution of protein identity values is similar across species pairs:
- Page 4, line 73. Which is the 23rd pair?
· Title and abstract
o Does the title clearly reflect the content of the article? [ ] Yes, [X] No (please explain), [ ] I don't know
I believe that the title could be improved to better describe the type of genomic changes that are assessed. Also, it should specify that the study is only considering metazoa.
o Does the abstract present the main findings of the study? [X] Yes, [ ] No (please explain), [ ] I don’t know
· Introduction
o Are the research questions/hypotheses/predictions clearly presented? [X] Yes, [ ] No (please explain), [ ] I don’t know
o Does the introduction build on relevant research in the field? [X] Yes, [] No (please explain), [ ] I don’t know
· Materials and methods
o Are the methods and analyses sufficiently detailed to allow replication by other researchers? [X] Yes, [ ] No (please explain), [ ] I don’t know
o Are the methods and statistical analyses appropriate and well described? [X] Yes, [ ] No (please explain), [ ] I don’t know
· Results
o In the case of negative results, is there a statistical power analysis (or an adequate Bayesian analysis or equivalence testing)? [ ] Yes, [ ] No (please explain), [ ] I don’t know
o Are the results described and interpreted correctly? [X] Yes, [ ] No (please explain), [ ] I don’t know
In general, however I have minor questions regarding the chromosome fusions and fissions.
· Discussion
o Have the authors appropriately emphasized the strengths and limitations of their study/theory/methods/argument? [X] Yes, [ ] No (please explain), [ ] I don’t know
o Are the conclusions adequately supported by the results (without overstating the implications of the findings)? [X] Yes, [ ] No (please explain), [ ] I don’t know
https://doi.org/10.24072/pci.genomics.100415.rev12
All in all, the authors have done an excellent job exploring the effects of speciation on genome structure. Congratulations on an excellent manuscript! I am looking forward to seeing this in print and I will definitely be adding it to my own citation library. I have a few comments below that I think would drastically improve the manuscript with very little work. These are personal opinions and I think the work is well-written and impactful as-is and other reviewers may not share these same thoughts.
Does the title clearly reflect the content of the article?
Yes. The title accurately reflect the content of the article. However, the author could stand to be a bit more specific as "genomic changes" is nonspecific and the title could likely improved slightly for higher impact.
Does the abstract present the main findings of the study?
Yes.
Are the methods and analyses sufficiently detailed to allow replication by other researchers?
Yes.
Are the methods and statistical analyses appropriate and well described?
Yes.
In the case of negative results, is there a statistical power analysis (or an adequate Bayesian analysis or equivalence testing)?
Yes.
Are the results described and interpreted correctly?
Yes.
Have the authors appropriately emphasized the strengths and limitations of their study/theory/methods/argument?
Mostly yes. My only concern is that the authors do not emphasize enough that they are working with species pairs that are diverged by highly variable amounts of time and are often not sister species. They do mention this on lines 34-45 on page 7 and 5-9 on page 6 but I think this needs to be even more thoroughly discussed at varying points in the manuscript since the observed processes might not directly be the result of speciation (which might be better observed in sister species pairs). The definition of a genus is as arbitrary as the definition of a species (which the authors do write about extensively in the introduction, maybe expand and describe how genera are also extremely arbitrary?) and some of the congeneric pairs are diverged by hundreds of millions of years (e.g. Daphnia) while others are diverged by only a few million (e.g. Drosophila) and are not sister (e.g. Daphnia pulex and Daphnia magna). The number of appropriate genomic resources available on NCBI specifically for sister species are scant (there are some good sister species references in other repositories but I strongly understand the desire to use NCBI over external databases) so I believe the authors have done an excellent job putting together an important paper with what is available. But I would specifically highlight that future work should explore the micro/macrosyntenic and identity differences between sister species since that could end up showing different results than seen here. I think all of my concerns would be alleviated by a few additional sentences that expand upon the limitations and potentially adding in a simple divergence time phylogeny (I don't actually want the authors to run the phylogenetic analysis, but instead put together a stylized figure that pulls from many different papers and depicts the divergence time and sister/non-sister relationships of the included taxa. Maybe even some kind of icon to depict the kind of speciation? e.g. allopatric vs sympatric).
Are the conclusions adequately supported by the results (without overstating the implications of the findings)?
Yes. The only change that I might make is to line 77 of page 7 where I would change species-pairs to sister species-pairs.
https://doi.org/10.24072/pci.genomics.100415.rev14