

Parasitic worms infect billions of animals worldwide. While parasitism is now considered a context-dependent relation along a symbiosis continuum, most of these parasitic worms, also known as helminths, can cause diseases that have a significant impact (Hopkins et al. 2017; Selzer, Epe 2021). When considering livestock animals, these impacts have a high economic cost, and therefore, prophylactic drugs are widely used (Selzer and Epe 2021). Consequently, drug resistance has become increasingly common across all parasites and concerns about drug effects on non-target organisms have been raised (de Souza and Guimarães 2022). This is why understanding the relationship between parasitic worms and their animal hosts and the diseases they cause at the genetic and molecular level is high on the agenda of parasitologists (Doyle 2022). The development of genomics resources plays a pivotal role in this agenda and is at the origin of Sallé and colleagues' article (2025).
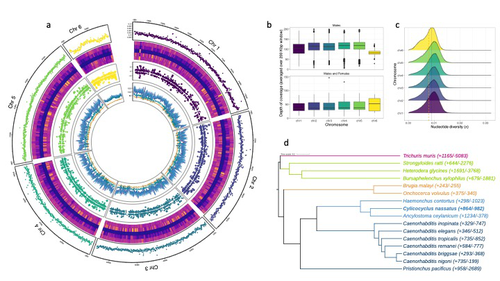
The most common intestinal parasites in equids are helminths of the cyathostomin nematode complex. These are the primary parasitic cause of death in young horses and also exhibit a reduced sensitivity to anthelmintic drugs. Therefore, Sallé and colleagues embarked on the arduous journey to build a reference annotated genome of the Cylicocylus nassatus nematode. They used cutting-edge molecular genetics methods to amplify and sequence the genome of a single individual and obtained chromosomal-level contiguity using Hi-C technology for six chromosomes and an assembly of 514.7 Mbp. Remarkably, transposable elements occupy more than half of the C. nassatus genome and may have led to an increase in genome size in this nematode. In parallel, the authors built a gene catalogue using transcriptomic data, reaching a BUSCO gene completion score of 94.1% with 22,718 protein-coding genes. They quantified allele frequencies based on the resequencing of nine populations, including an ancient Egyptian worm from the 19th century, indicating a recent loss of genetic diversity in European cyathostomin even if geographical sampling was limited. They also analysed transcriptomic differences between sexes and found differences linked with drug treatment. While there may be confounding effects due to global differences between sex that could explain this finding, these results will likely fuel future transcriptomic analyses investigating the response to antiparasitic drugs.
The Cylicocylus nassatus genome assembly obtained will be invaluable for studying nematode genome evolution and analysing the genetic and molecular basis of drug resistance in these parasites.
References
Doyle SR (2022) Improving helminth genome resources in the post-genomic era. Trends in Parasitology, 38, 831–840. https://doi.org/10.1016/j.pt.2022.06.002
Hopkins SR, Wojdak JM, Belden LK (2017) Defensive symbionts mediate host–parasite interactions at multiple scales. Trends in Parasitology, 33, 53–64. https://doi.org/10.1016/j.pt.2016.10.003
Sallé G, Courtot É, Cabau C, Parrinello H, Serreau D, Reigner F, Gesbert A, Jacquinot L, Lenhof O, Aimé A, Picandet V, Kuzmina T, Holovachov O, Bellaw J, Nielsen MK, Samson-Himmelstjerna G von, Valière S, Gislard M, Lluch J, Kuchly C, Klopp C (2024) Spatio-temporal diversity and genetic architecture of pyrantel resistance in Cylicocyclus nassatus, the most abundant horse parasite. bioRxiv, ver. 2 peer-reviewed and recommended by PCI Genomics https://doi.org/10.1101/2023.07.19.549683
Selzer PM, Epe C (2021) Antiparasitics in animal health: quo vadis? Trends in Parasitology, 37, 77–89. https://doi.org/10.1016/j.pt.2020.09.004
de Souza RB, Guimarães JR (2022) Effects of avermectins on the environment based on its toxicity to plants and soil invertebrates–a review. Water, Air, and Soil Pollution, 233, 259. https://doi.org/10.1007/s11270-022-05744-0
The reviewers have responded to all the concerns I raised and have edited their manuscript accordingly. I think the manuscript can be published in its current form.
DOI or URL of the preprint: https://doi.org/10.1101/2023.07.19.549683
Version of the preprint: 1
Dear recommender,
Please find attached our point-by-point response to the referees and the corresponding revised version.
Best wishes,
Guillaume Sallé
Dear Guillaume Salé and colleagues,
Your manuscript entitled "Spatio-temporal diversity and genetic architecture of pyrantel resistance in Cylicocyclus nassatus, the most abundant horse parasite" has been reviewed by two colleagues. Globally the reviews are of high quality and very positive, and find that your study has many merits, but there are a some criticisms that you need to adress before I can finally decide on this preprint recommendation.
With my best wishes,
Nicolas Pollet
The paper is an interesting read and makes a very valuable contribution to the equine parasitology research community. The authors are to be commended for taking a very methodical and careful approach to delivering a high-quality genome for a parasite species that has proved challenging in the past. Cyathostomins are not very tractable organisms with which to work and this manuscript reports a first success in provding a chromosomal level genome assembly. The work is very well done and is articulately described in the text. It provides interesting insights in cyathostomin biology and moves on the field significantly. The paper is generally well written and the authors provide convincing evidence for almost all of their conclusions. I have just one query for them to address:
My query centres around the section ‘Transcriptomic differences across sexes upon pyrantel exposure’. The authors propose that differences in the expression profiles between males and female may be due to developing eggs in the females or sex-specific differences in their response to pyrantel. However, there could be many reasons for the differences between the sexes, gastrointestinal nematode infections are not equal in terms of the number of male and female worms residing in the gut. Female worms are often more numerous and male worms can be polyandrous by nature. This raises the prospect of males being more motile as they move from mate to mate, which is one other way to explain differences between the sexes. There will likely be many other differences between the sexes. I remain unconvinced that the difference in their data is due to sex-dependent sensitivity as stated in lines 610-611 and would prefer to see some further discussion of these data.
Minor typos and text edits
1. Line 60 ‘on’ rather than ‘to’ per and livestock species
2. Line 62 ‘to’ the brink of distinction
3. Line 65 ‘anthelmintic’
4. Lines 70 and 72 ‘S. vulgaris’ and ‘C. nassatus’ should be in italics, if fact there are many examples throughout the text where parasite names are used and not italicised. They all need to be corrected.
5. Lines 71-73 use a comma rather than a hyphen
6. Line 75 ‘and’ zebras
7. Line 108 ‘decreased’
8. 129 ‘Ukraine to Kentucky’
9. Line 130-132 sentence needs reworking – what does ‘in that species’ refer to?
10. Line 317 ‘end-repaired is repeated’
11. Line 423 ‘Cylicostephanus’ spelling
12. Line 504 ‘The last century affected allele frequencies over known anthelmintic drug targets’ sounds a little clumsy. Do the authors mean ‘Allele frequencies of known anthelmintic targets has altered over the last century’? Or ‘Alteration in allele frequencies over the last century are found within genomic regions encoding anthelmintic drug targets? I think this title needs rewording.
13. Line 562 Figure 3 legend a.’ The number of private SNPs for a given isolate’
14. Line 585 (reference isolate – this mean STR?)
15. Table 1 legend ‘Bold names were covered by at least one? Decisive SNP….’
16. Figure 5 The legend and citation of this figure in the text is somewhat confusing and requires clarification. Some further explanation of the relationship between FECRT and allele frequency needs to be provided.
17. Line numbers now absent – First paragraph page 21. ‘can be reproduced in another organism’ – this sentence needs clarification as to what organisms this is referring to.
18. Line 9 page 22 ‘about twice the size’ or twice as large’
19. Paragraph 3 for ‘complexifies’ substitute ‘becomes more complex, as it remains to be determined….’?
The manuscript “Spatio-temporal diversity and genetic architecture of pyrantel resistance in Cylicocyclus nassatus, the most abundant horse parasite” by Sallé et al. presents a detailed study of the genome and genetic diversity of Cylicocyclus nassatus, the most prevalent species from the cyathostomin complex, a group of parasitic nematodes that infect horses and wild equids. The manuscript is the result of an impressive set of work: the authors have taken an important parasite from a clade of nematodes with practically no genomic data and have generated a very high-quality genome, a resequencing dataset, and sex-specific transcriptomes. They use these datasets to explore the genetic basis of pyrental resistance. The results are interesting and highlight several candidate loci that may play a role in pyrantel sensitivity. The manuscript will be of interest to those interested in parasite genetics and drug resistance in nematodes and flatworms. I commend the authors for making all data available on the relevant INSDC databases, which facilitated my review of their manuscript.
My only major comment is that the manuscript, in some places, suffers from an overly simplistic or one-sided interpretation of the results (which I detail below). In most cases, this doesn’t invalidate the major claims of the manuscript, but providing a more nuanced discussion of the results would really help readers understand the strengths and limitations of the study.
General comments
Although the reference genome is evidently of high quality, I found the presentation of the genome assembly results lacking detail in places, which led me to download the genome and annotation and calculate my own metrics. The following additions would help:
In my opinion, the authors’ analysis of gene family evolution adds very little to the manuscript:
I am not an expert on population genetic analyses, so other reviewers may be able to critically assess those sections better than I could, but I do have the following comments/questions:
The authors used a range of techniques and approaches to sequence the C. nassatus genome, but it was often unclear why they used the methods they did, and how the data were used during assembly. Given their genome sequencing/assembly methods are likely to be of interest to many readers, it would be great to be clearer in the methods and results about what they did:
The bias in sex ratios after pyrantel treatment is interesting. However, I am not fully convinced that the transcriptomic differences the authors identify are the result of sex-specific differences in response to pyrantel treatment:
Minor comments
The term ‘telomere-to-telomere’ refers to gap-free genome assembly (e.g. the new human reference genome, or the C. elegans reference genome). There are no telomere-to-telomere strongylid genomes. Better to use ‘chromosome-level’ or similar.
The Hi-C plot in Figure S1 appears to be truncated.
There doesn’t seem to be any information in the methods about how the C. nassatus genome was aligned to the H. contortus genome (although promer is noted in the Figure S3 caption). Also, using one-to-one orthologues, rather than promer, might reduce some of the noise seen in Figure S3.
It might just be me but I think it's better to write 19th century rather than XIXth.
The authors might want to define what they mean by ‘decisive SNP’.