

 based on reviews by 2 anonymous reviewers
based on reviews by 2 anonymous reviewers
The transition of cetaceans (whales, dolphins, and porpoises) from terrestrial to aquatic lifestyles is a striking example of natural selection driving major phenotypic changes (Figure 1). For instance, cetaceans have evolved the ability to withstand high pressure and to store oxygen for long periods, among other adaptations (Das et al. 2023). Many phenotypic changes, such as shifts in organ structure, have been well-characterized through fossils (Thewissen et al. 2009). Although such phenotypic transitions are now well understood, we have only a partial understanding of the underlying genetic mechanisms. Scanning for signatures of adaptation in genes related to phenotypes of interest is one approach to better understand these mechanisms. This was the focus of Uribe and colleagues’ (2024) work, who tested for such signatures across cetacean protein-coding genes.

Figure 1: The skeletons of Ambulocetus (an early whale; top) and Pakicetus (the earliest known cetacean, which lived about 50 million years ago; bottom). Copyright: J. G. M. Thewissen. Displayed here with permission from the copyright holder.
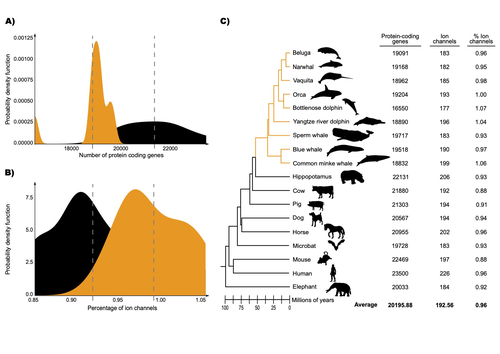
The authors were specifically interested in investigating the evolution of ion channels, as these proteins play fundamental roles in physiological processes. An important aspect of their work was to develop a bioinformatic pipeline to identify orthologous ion channel genes across a set of genomes. After applying their bioinformatic workflow to 18 mammalian species (including nine cetaceans), they conducted tests to find out whether these genes showed signatures of positive selection in the cetacean lineage. For many ion channel genes, elevated ratios of non-synonymous to synonymous substitution rates were detected (for at least a subset of sites, and not necessarily the entire coding region of the genes). The genes concerned were enriched for several functions, including heart and nervous system-related phenotypes.
One top gene hit among the putatively selected genes was SCN5A, which encodes a sodium channel expressed in the heart. Interestingly, the authors noted a specific amino acid replacement, which is associated with sensitivity to the toxin tetrodotoxin in other lineages. This substitution appears to have occurred in the common ancestor of toothed whales, and then was reversed in the ancestor of bottlenose dolphins. The authors describe known bottlenose dolphin interactions with toxin-producing pufferfish that could result in high tetrodotoxin exposure, and thus perhaps higher selection for tetrodotoxin resistance. Although this observation is intriguing, the authors emphasize it requires experimental confirmation.
The authors also recapitulated the previously described observation (Yim et al. 2014; Huelsmann et al. 2019) that cetaceans have fewer protein-coding genes compared to terrestrial mammals, on average. This signal has previously been hypothesized to partially reflect adaptive gene loss. For example, specific gene loss events likely decreased the risk of developing blood clots while diving (Huelsmann et al. 2019). Uribe and colleagues also considered overall gene turnover rate, which encompasses gene copy number variation across lineages, and found the cetacean gene turnover rate to be three times higher than that of terrestrial mammals. Finally, they found that cetaceans have a higher proportion of ion channel genes (relative to all protein-coding genes in a genome) compared to terrestrial mammals.
Similar investigations of the relative non-synonymous to synonymous substitution rates across cetacean and terrestrial mammal orthologs have been conducted previously, but these have primarily focused on dolphins as the sole cetacean representative (McGowen et al. 2012; Nery et al. 2013; Sun et al. 2013). These projects have also been conducted across a large proportion of orthologous genes, rather than a subset with a particular function. Performing proteome-wide investigations can be valuable in that they summarize the genome-wide signal, but can suffer from a high multiple testing burden. More generally, investigating a more targeted question, such as the extent of positive selection acting on ion channels in this case, or on genes potentially linked to cetaceans’ increased brain sizes (McGowen et al. 2011) or hypoxia tolerance (Tian et al. 2016), can be easier to interpret, as opposed to summarizing broader signals. However, these smaller-scale studies can also experience a high multiple testing burden, especially as similar tests are conducted across numerous studies, which often is not accounted for (Ioannidis 2005). In addition, integrating signals across the entire genome will ultimately be needed given that many genetic changes undoubtedly underlie cetaceans’ phenotypic diversification. As highlighted by the fact that past genome-wide analyses have produced some differing biological interpretations (McGowen et al. 2012; Nery et al. 2013; Sun et al. 2013), this is not a trivial undertaking.
Nonetheless, the work performed in this preprint, and in related research, is valuable for (at least) three reasons. First, although it is a challenging task, a better understanding of the genetic basis of cetacean phenotypes could have benefits for many aspects of cetacean biology, including conservation efforts. In addition, the remarkable phenotypic shifts in cetaceans make the question of what genetic mechanisms underlie these changes intrinsically interesting to a wide audience. Last, since the cetacean fossil record is especially well-documented (Thewissen et al. 2009), cetaceans represent an appealing system to validate and further develop statistical methods for inferring adaptation from genetic data. Uribe and colleagues’ (2024) analyses provide useful insights relevant to each of these points, and have generated intriguing hypotheses for further investigation.
References
Das K, Sköld H, Lorenz A, Parmentier E (2023) Who are the marine mammals? In: “Marine Mammals: A Deep Dive into the World of Science”. Brennecke D, Knickmeier K, Pawliczka I, Siebert U, Wahlberg, M (editors). Springer, Cham. p. 1–14. https://doi.org/10.1007/978-3-031-06836-2_1
Huelsmann M, Hecker N, Springer MS, Gatesy J, Sharma V, Hiller M (2019) Genes lost during the transition from land to water in cetaceans highlight genomic changes associated with aquatic adaptations. Science Advances, 5, eaaw6671. https://doi.org/10.1126/sciadv.aaw6671
Ioannidis JPA (2005) Why most published research findings are false. PLOS Medicine, 2, e124. https://doi.org/10.1371/journal.pmed.0020124
McGowen MR, Montgomery SH, Clark C, Gatesy J (2011) Phylogeny and adaptive evolution of the brain-development gene microcephalin (MCPH1) in cetaceans. BMC Evolutionary Biology, 11, 98. https://doi.org/10.1186/1471-2148-11-98
McGowen MR, Grossman LI, Wildman DE (2012) Dolphin genome provides evidence for adaptive evolution of nervous system genes and a molecular rate slowdown. Proceedings of the Royal Society B: Biological Sciences, 279, 3643–3651. https://doi.org/10.1098/rspb.2012.0869
Nery MF, González DJ, Opazo JC (2013) How to make a dolphin: molecular signature of positive selection in cetacean genome. PLOS ONE, 8, e65491. https://doi.org/10.1371/journal.pone.0065491
Sun YB, Zhou WP, Liu HQ, Irwin DM, Shen YY, Zhang YP (2013) Genome-wide scans for candidate genes involved in the aquatic adaptation of dolphins. Genome Biology and Evolution, 5, 130–139. https://doi.org/10.1093/gbe/evs123
Tian R, Wang Z, Niu X, Zhou K, Xu S, Yang G (2016) Evolutionary genetics of hypoxia tolerance in cetaceans during diving. Genome Biology and Evolution, 8, 827–839. https://doi.org/10.1093/gbe/evw037
Thewissen JGM, Cooper LN, George JC, Bajpai (2009) From land to water: the origin of whales, dolphins, and porpoises. Evolution: Education and Outreach, 2, 272–288. https://doi.org/10.1007/s12052-009-0135-2
Uribe C, Nery M, Zavala K, Mardones G, Riadi G, Opazo J (2024) Evolution of ion channels in cetaceans: A natural experiment in the tree of life. bioRxiv, ver. 8 peer-reviewed and recommended by Peer Community in Genomics. https://doi.org/10.1101/2023.06.15.545160
Yim HS, Cho YS, Guang X, Kang SG, Jeong JY, Cha SS, Oh HM, Lee JH, Yang EC, Kwon KK, et al. (2014) Minke whale genome and aquatic adaptation in cetaceans. Nature Genetics, 46, 88–92. https://doi.org/10.1038/ng.2835
DOI or URL of the preprint: https://www.biorxiv.org/content/10.1101/2023.06.15.545160v7
Version of the preprint: 7
Dear Gavin, thank you for your last suggestion. The title of the manuscript was modified according to your suggestion.
All the best
Juan
 , posted 22 Apr 2024, validated 22 Apr 2024
, posted 22 Apr 2024, validated 22 Apr 2024Hi Dr. Opazo and colleagues,
Thanks for making those corrections and for replying to my comments. I am now happy to recommend your manuscript, which should be online soon.
One final point before I do so, however -- I just noticed that the wording of your title is actually grammatically incorrect: "Evolution of ion channels in cetaceans a natural experiment in the tree of life". It needs to be fixed to be one of these options (or something similar):
I suggest the first option (just adding a colon, and capitalizing "A").
Sincerely,
Gavin Douglas
DOI or URL of the preprint: https://www.biorxiv.org/content/10.1101/2023.06.15.545160v4
Version of the preprint: 4
 , posted 07 Apr 2024, validated 10 Apr 2024
, posted 07 Apr 2024, validated 10 Apr 2024Hi Dr. Opazo and colleagues,
I have read through your updated manuscript and responses. I find it has greatly improved, and the reviewers’ comments have been adequately addressed. However, prior to recommending your work I do have a few final comments that I would like to see addressed.
My major comment is regarding the functional enrichment analysis. You clarified that only ion channel genes were used for this analysis, but readers will likely find this somewhat confusing as they will likely expect the analysis to have been conducted against the background of all genes, rather than just ion channel genes. This distinction means that although the ion channel genes/functions identified through this analysis are indeed the best candidates for positive selection among ion channel genes, they are not necessarily the strongest candidates of positive selection across all cetacean genes. Explicitly clarifying this point would be important, as otherwise it could be misleading about how important ion channel genes may have been for the aquatic transition.
On a related point, although Figure 3 provides a clear high-level overview, there are insufficient details given to help the readers evaluate the overall results. Readers need to dig into the supplementary tables to see specific details, which are tables of P-values for significant categories. Although significance tests are important, a report on the effect sizes of the tested categories would be more relevant, and would help readers evaluate the work. I strongly recommend that a figure or small table be (re-)added to the main text to provide more specific details to help readers evaluate this analysis (ideally that would contain effect sizes, e.g., of odd’s ratios of how enriched a certain gene / function is among positively selected genes, or the actual count breakdown). The original Table 1 was better, but without effect sizes (or at least the numbers of genes in each category vs not in that category) it was not very informative.
Last, in the enrichment analysis methods section, the authors describe identifying significant hits with an “adjusted probability value of less than 0.01” and where “The adjusted probability is calculated from the resulting list of categories with raw p-values equal to or lower than 0.05 through the procedure of [FDR]”. This makes it sound like FDR correction was run based on the set of raw P-values below 0.05, which would be incorrect (this correction would need to be run on the set of raw P-values for all tests, not just those considered significant). If I am interpreting this description correctly, then the authors would need to change their correction approach and the enrichment results would need to be re-analyzed after appropriately applying multiple-test correction. Please clarify what was done exactly and whether I am misunderstanding what correction was performed.
Other comments
The authors use the phrase “conquest of the aquatic environment” in numerous places. I strongly suggest that they use phrasing like “aquatic transition” instead, at least in most cases.
It would help readers who are non-experts in the cetacean field if the crown and stem clades were indicated on the phylogenetic tree. Please add this to Figure 2.
I suggest you add a README file to your GitHub repository to describe the scripts (or at least the subfolders), and you could unzip the scripts so they can be explored online as well. Also, please add the supplementary tables to a more permanent location, since GitHub repos can be taken down over time. For example, a Zenodo repository would work better for this purpose.
In the abstract, ion channels are first brought up in an abrupt way: “Ion channels are a crucial component of the cellular machinery for the proper physiological functioning of all living species.” My understanding is that the authors specifically are interested in ion channels because they hypothesize that they might have particularly been under selection for physiological changes during the aquatic transition. Bringing up ion channels in this context would be a lot clearer, as at the moment it is not clear why you focused on them specifically (in the abstract).
More details on where the phylogenetic tree was taken from are needed (this is reported in a figure legend, but should be in the methods itself). If the tree generation required some manual steps, it would be good to include these in your GitHub repo. Otherwise please make it clear where the file was downloaded from.
In the methods, I found this sentence a little vague: “Both analyses take into account the sister group relationships of the included species”. It would be good to clarify what exactly is meant, e.g., do you mean that the phylogenetic similarity of species was taken into account for these analyses? Or something specifically about sister species?
The dN/dS null models are described as only permitting sites with omega < 1, but to my understanding these models permit sites with omega <= 1 (e.g., see Table 1 of this paper: https://doi.org/10.1093/molbev/msad041).
Please include a mention of the Creative Common license that the silhouette images downloaded from PhyloPic are under (and include a link to the license if that is required under the re-use terms).
The authors mention: “Among the associated genes related to heart physiology in cetaceans … the Sodium Voltage-Gated Channel Alpha Subunit 5 (SCN5A) is the most frequent.” I think it would be more correct to state that homologs to this gene were identified, or use genes (plural), rather than “the gene”. My thinking is that there are presumably multiple such genes per genome if it was the most frequent hit, so clearing that up grammatically would be good.
Nav1.5 is defined in regards to SCN5A, but it would be good to quickly describe the relationship with Nav1.7 too, as this wasn’t clear to me upon re-reading.
Related to the above, I found the paragraph starting with "Although the NaV1.5 and NaV1.7 sodium channels have similar selectivity filters” (page 13) confusing, as it began as a comparison between Nav1.5 and Nav1.7 and then changes to contrasting human and cetacean Nav1.5. Please rephrase this sentence. Introducing Nav1.7 vs Nav1.5 would be a good starting point.
“Frogs” are mentioned as marine animals at the top of page 13. Please correct this.
Please note -- I have corrected a few typos and made some minor suggestions in the attached Word document (with tracked changes).
Download recommender's annotations
DOI or URL of the preprint: https://www.biorxiv.org/content/10.1101/2023.06.15.545160v4
Version of the preprint: 4
 , posted 09 Feb 2024, validated 09 Feb 2024
, posted 09 Feb 2024, validated 09 Feb 2024I thank the authors for their resubmission, but the response to my own comments was missing. Please address the comments in the PDF I provided previously in addition to the two reviewers.
Thanks,
Gavin Douglas
DOI or URL of the preprint: https://doi.org/10.1101/2023.06.15.545160
Version of the preprint: 2
 , posted 20 Sep 2023, validated 21 Sep 2023
, posted 20 Sep 2023, validated 21 Sep 2023Please see my decision and comments in the attached PDF. I would be happy to also provide this file as a Word document if that is preferred when formulating your response.
Download recommender's annotationsThis study employs a bioinformatics pipeline to investigate the evolutionary dynamics of ion channels in cetaceans. The findings reveal a reduction in the repertoire of ion channels in cetaceans compared to their terrestrial mammalian counterparts. Notably, the NaV1.5 ion channel in most toothed whales exhibits specific amino acid variations deemed pathological in humans. Particularly, a significant proportion of these whales possess a tyrosine residue at a precise position within the NaV1.5 channel, potentially rendering them more susceptible to certain toxins. These discoveries offer profound insights into the mechanisms underpinning cetacean adaptations to their aquatic habitat. This research not only presents intriguing implications but also holds substantial scientific significance. The study encompasses a variety of functionalities related to ion channels, including cardiac and skeletal muscle contraction, echolocation, and polycystic kidney syndrome. However, experimental validation of these bioinformatic analyzes is necessary and requires in-depth investigation of the specific functions of ion channels.
https://doi.org/10.24072/pci.genomics.100256.rev12