

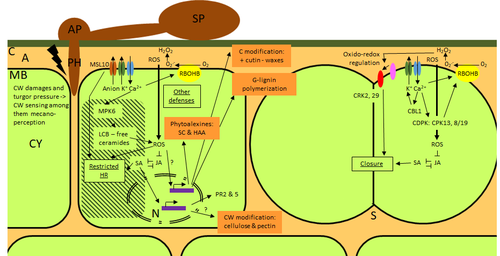
Nonhost resistance is a common form of disease resistance exhibited by plants against microorganisms that are pathogenic to other plant species [1]. Apples and pears are two closely related species belonging to Rosaceae family, both affected by scab disease caused by fungal pathogens in the Venturia genus. These pathogens appear to be highly host-specific. While apples are nonhosts for Venturia pyrina, pears are nonhosts for Venturia inaequalis. To date, the molecular bases of scab nonhost resistance in apple and pear have not been elucidated.
This preprint by Vergne, et al (2022) [2] analyzed nonhost resistance symptoms in apple/V. pyrina and pear/V. inaequalis interactions as well as their transcriptomic responses. Interestingly, the author demonstrated that the nonhost apple/V. pyrina interaction was almost symptomless while hypersensitive reactions were observed for pear/V. inaequalis interaction. The transcriptomic analyses also revealed a number of differentially expressed genes (DEGs) that corresponded to the severity of the interactions, with very few DEGs observed during the apple/V. pyrina interaction and a much higher number of DEGs during the pear/V. inaequalis interaction.
This type of reciprocal host-pathogen interaction study is valuable in gaining new insights into how plants interact with microorganisms that are potential pathogens in related species. A few processes appeared to be involved in the pear resistance against the nonhost pathogen V. inaequalis at the transcriptomic level, such as stomata closure, modification of cell wall and production of secondary metabolites as well as phenylpropanoids. Based on the transcriptomics changes during the nonhost interaction, the author compared the responses to those of host-pathogen interactions and revealed some interesting findings. They proposed a series of cascading effects in pear induced by the presence of V. inaequalis, which I believe helps shed some light on the basic mechanism for nonhost resistance.
I am recommending this study because it provides valuable information that will strengthen our understanding of nonhost resistance in the Rosaceae family and other plant species. The knowledge gained here may be applied to genetically engineer plants for a broader resistance against a number of pathogens in the future.
References
1. Senthil-Kumar M, Mysore KS (2013) Nonhost Resistance Against Bacterial Pathogens: Retrospectives and Prospects. Annual Review of Phytopathology, 51, 407–427. https://doi.org/10.1146/annurev-phyto-082712-102319
2. Vergne E, Chevreau E, Ravon E, Gaillard S, Pelletier S, Bahut M, Perchepied L (2022) Phenotypic and transcriptomic analyses reveal major differences between apple and pear scab nonhost resistance. bioRxiv, 2021.06.01.446506, ver. 4 peer-reviewed and recommended by Peer Community in Genomics. https://doi.org/10.1101/2021.06.01.446506
The authors’ changes to make their DEG results more quantitative has greatly improved their manuscript. However, I do have some minor comments.
Regarding Figure 2 – this is a great addition. I would change the second column title be something like “Odd’s ratio vs background” and then explain precisely what you mean in the legend. Currently the column title is not strictly correct, as it is the relative proportions of the category in the DEGs and background that are being compared, not the raw frequencies. This should be explicitly explained in the methods, as the use of “ratio” in the text is a bit ambiguous. Also, it is standard to plot odd ratios plots on a log-scale (usually log2), so that the magnitude of over and under enrichments vs baseline can be compared more clearly. An x-axis label should also be added. It’s not clear to me what the “input set freq.” values refer to, and they should either be removed or explained if they are pertinent. Finally, note that “Frequence” should be “Frequency” and “Fonctional” should be “Functional”.
In Table 2 – “% upregulated DEGs” should be changed to “% DEGs upregulated” (and likewise for the downregulated line) for clarity
The results subsection titles are fine, but should be made bold or otherwise different from rest of text to. Also, on L126 – should be “symptom analysis” rather than “symptoms analyze”. Similarly, on L147, “analyze” should be “analysis”.
The subsection title changes in the discussion make the meaning much clearer: they now appropriately make the findings seem like hypotheses rather than clear evidence.
I noticed on bioRxiv that a bot identified that you cite a retracted paper. You can see the Twitter post here: https://twitter.com/sciterefcheck/status/1410812068412891146?s=20&t=RDkN6IYvOV4qeDTfwXuEFA And this is the retracted paper you cite, which should be removed: https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0242041. This citation was used to support a minor point in the text, for which I think the authors can find alternative support or simply remove.
DOI or URL of the preprint: https://doi.org/10.1101/2021.06.01.446506
Version of the preprint: 2
Dear Authors,
The reviewers have looked at the manuscript, and two of them provided extensive and valuable feedbacks. Please take a look at their comments below and revise your manuscript accordingly. Please pay special attention to the comments on the enrichment test as I also think it is important to carry out such test. In addition, please make sure that the hypothesis or anything that is not supported by the evidence provided in this study is written in a way that conveys the "hypothetical" nature of it (the language used should not be too ascertain). Please let us know if you require an extension on the revision of the paper.
Regards,
Wirulda Pootakham
Vergne and colleagues present a comprehensive investigation into the transcriptomic changes following pear scab nonhost resistance to a pathogen. The authors clearly have detailed knowledge of the expected (and possible) immune responses in pears and apples and I think the description of these pathways alone would be useful for researchers in this area.
However, this paper suffers from a severe flaw: it is largely qualitative after the initial differential expression testing. Specifically, the authors devote most of the manuscript to introducing various expected immune and physical pathways and phenotypes and then mention how many genes related to those traits were differentially expressed. They then dig into specifically what those genes are. The problem with this approach is that it is not at all convincing that genes overall related to the phenotypes of interest are differentially expressed. In other words, a few genes in each of these pathways/phenotypes of interest may be differentially expressed, but possibly a similar number would be found if genes were totally randomly sampled. Standard enrichment tests (e.g., Fisher’s exact tests) are needed to support the authors' claims that these traits are specifically enriched or depleted for differentially expressed genes. Otherwise it is very possible that the authors could be interpreting noise in their data – the reader currently has little means to evaluate that.
This manuscript is also quite unique in that it has a very long results/discussion section, which tends to largely be a discussion of the predicted gene functions and other results in the literature. I think it would be much clearer if the authors elected instead to include a brief results section where they present the results of enrichment tests such as I alluded to above. They should also provide specific odd’s ratios and p-values where relevant as well. A quick summary of the results in this way would make the paper much easier to interpret. The authors could then expand specific genes within the enriched pathways/phenotypes and make connections to other literature. As currently the paper is 90% discussion, I think keeping it in the current format would make it very difficult for the reader to disentangle exactly what the data presented here supports.
Other major comments
In general, the authors are too strong in how they interpret the RNA-seq data. For instance, at L190-191 I disagree that the RNA-seq data is clear evidence that “some JA seems to be produced, but [is] rapidly converted [into] inactive compounds…” The data is consistent with this hypothesis, but I think the language should be toned down as you are not actually measuring JA levels here. Similar issues are at play in the conclusion paragraphs of each discussion section: the authors should make it clear that they are indirectly inferring the levels of calcium influx, HR, etc. Otherwise readers would get the wrong impression for how much confidence they should put in all of these very specific inferences. E..g, L263-266: the authors speak about calcium influx and development of specific stomatal closure pre-invasive defense as though it were experimentally shown, but there is no direct evidence for this. There are many similar examples as well, e.g., L325:327, L409:410, etc.
The same problem is at play with the subsection titles. For instance, the authors do not have direct evidence that the cell wall carbohydrates content and cuticle content are altered”, only that some genes likely involved in those processes are differentially expressed. This is a very important distinction.
The introduction should be lengthened to discuss some more prior results. Currently it feels unbalanced as there is a very long results and discussion section, but concise introduction. I think this it would be especially important to introduce citation 11 more, which comes up numerous times. It would be helpful if the authors made the differences between this paper and their own clear from the offset (this can be inferred from the last sentence of the intro with a careful reading, but it would be better if this was explicit).
I don’t find Figure 2 very informative – the key question I have is what is the background proportion of all of those categories? I think rather than having the percent of total genes on the x-axis, it could just be the percent of all significant hits, and you could show the bars for the percents of background genes in all categories as well, to make it easier to evaluate whether any categories are particularly common (or depleted). This is of course related to my key critique of the manuscript as well.
Exact sample sizes and replicate structure should be more clearly explained and brought up at the beginning of the results as well.
Custom code used for project should be made available through an online repository, e.g., on GitHub. It is not sufficient to point to R and AnaDiff. Also, I don’t think the authors explicitly mention AnaDiff in the manuscript itself – details on the actual statistical test for differential expression testing are needed.
Minor
First line of each paragraph (except for the first paragraph of a section, which is optional) should be indented.
L19 (and elsewhere): is being “a nonhost” the correct term? Or should it be they “have nonhost resistance to …”
L49 – Add “.” after “et al”
L80 – Make it clear that Gala and Conference are an apple and pear cultivar, respectively, upon first mention
Table 1: Should add the description of each class as another column of table, so thjat the reader doesn’t have to keep looking back and forth
Table 2: Perhaps change “without TAIR name” to “without Arabidopsis homolog”?
Should be space between number “hpi”, e.g., 72hpi should be 72 hpi
L143 – “theses” should be “these”
L159 – remove “basically”
L159 – “DEGS” should be “DEGs” and “have been tested” should be “were tested”
Figure 2: decimal places on x-axis should be period
L161: “weak” should be “low”
L168 – Use active voice when describing results found in this work, or make it clear if you’re referring to a different paper
L191 – add “is” before “rapidly” and “into” instead of “in”
L270: “i. e.” should be “, i.e.,”
L316: add “the” before JA
L333-L336 – Re-word, this sentence is very hard to follow.
L606: Replace “As far as we know” with “To our knowledge”
Vergne and co-workers have here deciphered and compared the mechanisms underlying NHR of apple and pear using phenotypic and transcriptomic analyses. They have shown that the resistance differed in terms of phenotypic expression of the resistance, and that the difference was also mirrored by the gene expression underlying resistance, with DEGs being consistent with the phenotypic expression of the resistance in both plant species.
I particularly appreciated that the transcriptomic data were thoroughly explored and that authors have illustrated their findings through well-designed figures.
Nonhost resistance is increasingly considered as a promising field of research to identify sustainable disease-control methods with a low environmental impact, and the authors have provided a high-quality analysis of their data. Considering that, this paper brings valuable information for the community and I fully support the publication of this article in PCI Genomics once comments have been taken in consideration.
Major comments:
- even though very few genes are DEG in the apple / V. pyrina interaction, could you provide the list of these genes in suppl. data, with as much info regarding these genes as you have?
- lines 82-90: could you indicate in the text the number of interactions tested for each pathosystem? Even though a few infections of pears with V. inequalis gave rise to HR or resistance symptoms, most interactions (90%) were symptomless. It was 100% for the apple X V. pyrina. Could the symptoms observed, as they are not on all interactions, be due to an environmental effect? Also, to be cautious, I would mitigate the statement that the interaction is NH type II, just by adding "seems" or "most interactions were asymptomatic except for X interactions, we are hypothesizing that it is type II". This is also confusing because a “small scale HR” is observed in both cases (lines 104-105). Moreover, in both cases, growing of the hyphae seems to be very limited (from figure 1), although the outcome of the interaction is not the same, could you comment on that? For the pear x V. inequalis interaction, it could be the same as for the “rare HR-like reactions” on apple x V. pyrina (lines 108-109). Please comment and clarify. I think it would be sound not to conclude here on the type of HR but to use the transcriptomic analysis to validate the hypothesis.
- Figure 1: could you add the same visualization of the infection of apple with V. inequalis and pear with V. pyrina as a control, for non-familiar people.
- lines 117-119: justify the time points used for the transcriptomic analysis especially that later, 6 dpi are mentioned (line 146): do these timepoints correspond to a particular stage in the apple and / or pear infection by their respective adaptive pathogen? and are the inoculations made on detached leaf (cause possible leaf ageing is mentioned line 122)?
- lines 120-122: this sentence is difficult to follow, could you rephrase to clarify, for instance, remove “not to the infection but” and “which” should be used instead of “whose”.
- lines 149-150: have you compared these data to the infection of pear by V. pyrina or apple by V. inequalis (i.e type of gene deregulated due to infection by the adapted pathogen)? Authors may have done later in the paper.
- part “calcium influx and ROS…”: could you indicate which pathosystem this data corresponds to? And if it changes throughout this part? It was unclear at the first reading.
-line 476: “but further functional analyses…” could you indicate which analyses could be set up to conclude?
- lines 707-708: transposable elements were represented on the array, but no results were mentioned concerned these elements, could you provide the results?
-lines 798-710: were all genes of V. inaequalis represented on the microarray? Why were the data regarding these genes not presented?
Minor comments:
- in the text, sometimes “Malus x domestica”, sometimes “Malus domestica”, homogenize.
- line 80: add that leaf is inoculated even though it is mentioned in the methods.
- Figure 1: images extend beyond the frame and are not aligned. “V. pyrina”: the "V" must be italicized
- Table 1, line 97: a parenthesis is missing, add the number of interactions assessed in parenthesis.
- line 119-120: you can remove “kinetic”, “experimental design” is sufficient.
- line 125: to be consistent, change “differently” to “differentially”.
- table 2: to be consistent, add or remove “of” for the number of genes deregulated. Add the meaning of “TAIR”.
- table S1: header of columns G and H to be checked. Title should be “… BLAST analysis”.
- line 147: “a later” rather than “longer” should be written.
- line 154: “at both time points of the experiment” rather than “of the kinetics”.
- homegenise “up-regulated” or “upregulated”, same for down regulated throughout the paper.
- figure 2: replace “,” by “.”
- line 159: remove “basically”
- line 164: change “24phi” to “24hpi”
- line 165: I would conclude this part with a biological conclusion by moving the validation of the microarray data earlier in the text and finishing on the functional categories identified.
-line 170: change “that is” to “corresponding to”
-line 201: “…only two of the previously activated ones”?
- line 209: what do you mean with “SA accumulation was also rather mixed”?
- throughout the text, write “nonhost” rather than “non-host”, both are used and this needs to be consistent.
- line 312: change “hypersensitive reaction” to “HR”.
- line 336: “plant cell wall interactions, xxx”. Remove the coma.
- line 342: “xxx Table 3 xxx”
- Table 4: please, add the legend (for “*”) below the table
- line 433: change “hemi biotrophic” to “hemibiotrophic”
- line 434: change “strong induction” to “Strong induction…”
- lines 456-459: the “s” is missing for the verbs conjugated to the the 3rd person sing.
- line 512: “no known function” to “unpredicted function”?
-line 642: “in vitro” to be italicized