

Biological systems depend on finely tuned interactions of their components. Thus, regulating these components is critical for the system's functionality. In prokaryotic cells, riboswitches are regulatory elements controlling transcription or translation. Riboswitches are RNA molecules that are usually located in the 5′-untranslated region of protein-coding genes. They generate secondary structures leading to the regulation of the expression of the downstream protein-coding gene (Kavita and Breaker, 2022). Riboswitches are very versatile and can bind a wide range of small molecules; in many cases, these are metabolic byproducts from the gene’s enzymatic or signaling pathway. Their versatility and abundance in many species make them attractive for synthetic biological circuits. One class that has been drawing the attention of synthetic biologists is toehold switches (Ekdahl et al., 2022; Green et al., 2014). These are single-stranded RNA molecules harboring the necessary elements for translation initiation of the downstream gene: a ribosome-binding site and a start codon. Conformation change of toehold switches is triggered by an RNA molecule, which enables translation.
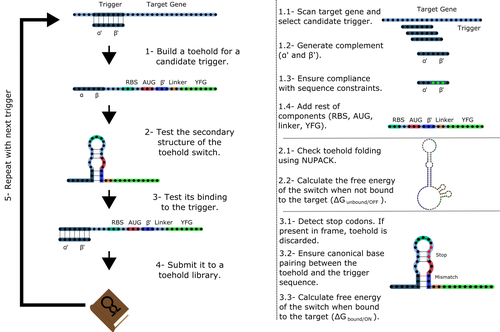
To exploit the most out of toehold switches, automation of their design would be highly advantageous. Cisneros and colleagues (Cisneros et al., 2022) developed a tool, “Toeholder”, that automates the design of toehold switches and performs in silico tests to select switch candidates for a target gene. Toeholder is an open-source tool that provides a comprehensive and automated workflow for the design of toehold switches. While web tools have been developed for designing toehold switches (To et al., 2018), Toeholder represents an intriguing approach to engineering biological systems by coupling synthetic biology with computational biology. Using molecular dynamics simulations, it identified the positions in the toehold switch where hydrogen bonds fluctuate the most. Identifying these regions holds great potential for modifications when refining the design of the riboswitches. To be effective, toehold switches should provide a strong ON signal and a weak OFF signal in the presence or the absence of a target, respectively. Toeholder nicely ranks the candidate toehold switches based on experimental evidence that correlates with toehold performance (based on good ON/OFF ratios).
Riboswitches are highly appealing for a broad range of applications, including pharmaceutical and medical purposes (Blount and Breaker, 2006; Giarimoglou et al., 2022; Tickner and Farzan, 2021), thanks to their adaptability and inexpensiveness. The Toeholder tool developed by Cisneros and colleagues is expected to promote the implementation of toehold switches into these various applications.
References
Blount KF, Breaker RR (2006) Riboswitches as antibacterial drug targets. Nature Biotechnology, 24, 1558–1564. https://doi.org/10.1038/nbt1268
Cisneros AF, Rouleau FD, Bautista C, Lemieux P, Dumont-Leblond N, ULaval 2019 T iGEM (2022) Toeholder: a Software for Automated Design and In Silico Validation of Toehold Riboswitches. bioRxiv, 2021.11.09.467922, ver. 3 peer-reviewed and recommended by Peer Community in Genomics. https://doi.org/10.1101/2021.11.09.467922
Ekdahl AM, Rojano-Nisimura AM, Contreras LM (2022) Engineering Toehold-Mediated Switches for Native RNA Detection and Regulation in Bacteria. Journal of Molecular Biology, 434, 167689. https://doi.org/10.1016/j.jmb.2022.167689
Giarimoglou N, Kouvela A, Maniatis A, Papakyriakou A, Zhang J, Stamatopoulou V, Stathopoulos C (2022) A Riboswitch-Driven Era of New Antibacterials. Antibiotics, 11, 1243. https://doi.org/10.3390/antibiotics11091243
Green AA, Silver PA, Collins JJ, Yin P (2014) Toehold Switches: De-Novo-Designed Regulators of Gene Expression. Cell, 159, 925–939. https://doi.org/10.1016/j.cell.2014.10.002
Kavita K, Breaker RR (2022) Discovering riboswitches: the past and the future. Trends in Biochemical Sciences. https://doi.org/10.1016/j.tibs.2022.08.009
Tickner ZJ, Farzan M (2021) Riboswitches for Controlled Expression of Therapeutic Transgenes Delivered by Adeno-Associated Viral Vectors. Pharmaceuticals, 14, 554. https://doi.org/10.3390/ph14060554
To AC-Y, Chu DH-T, Wang AR, Li FC-Y, Chiu AW-O, Gao DY, Choi CHJ, Kong S-K, Chan T-F, Chan K-M, Yip KY (2018) A comprehensive web tool for toehold switch design. Bioinformatics, 34, 2862–2864. https://doi.org/10.1093/bioinformatics/bty216
DOI or URL of the preprint: https://doi.org/10.1101/2021.11.09.467922
Version of the preprint: 2
Answer to reviewers (Round #2) :
Dear François,
Thank you for resubmitting your manuscript entitled "Toeholder: a Software for Automated Design and In Silico Validation of Toehold Riboswitches". You have successfully addressed all the comments from the reviewers. However, before we recommend on this manuscript there are minor revisions that are needed.
- Figure 2: The text is smaller in comparison to the other figures. Please enlarge it.
Response: Main text was standardized to 18pt in figure 2.
- Please consider dividing the "4.2 Toeholder conception and validation" subtitle into two subtitles.
Response: 4.2 was subdivided into 4.2 Toeholder conception and 4.3 Toeholder validation.
- 7.Data availability: please add the DOIs of your data, scripts and code in this section.
Response: DOIs were added to the section.
- Please add a section "10. Supplementary material" with the DOI/URL to your supplementary files (Supplementary video 1).
Response: Section 10 was created and DOI was added for Supplementary video 1.
On behalf of all authors on this paper, thank you very much for you feedback and thorough review of this paper! We enjoyed the process of submitting to PCI, and look forward to future collaborations.
Francois and Angel
Dear François,
Thank you for resubmitting your manuscript entitled "Toeholder: a Software for Automated Design and In Silico Validation of Toehold Riboswitches". You have successfully addressed all the comments from the reviewers. However, before we recommend on this manuscript there are minor revisions that are needed.
- Figure 2: The text is smaller in comparison to the other figures. Please enlarge it.
- Please consider dividing the "4.2 Toeholder conception and validation" subtitle into two subtitles.
- 7.Data availability: please add the DOIs of your data, scripts and code in this section.
- Please add a section "10. Supplementary material" with the DOI/URL to your supplementary files (Supplementary video 1).
Best regards,
Sahar
DOI or URL of the preprint: https://doi.org/10.1101/2021.11.09.467922
Version of the preprint: 1
Dear Dr. Cisneros,
Thank you for the opportunity to consider your manuscript at PCI Genomics. The two reviewers found the work interesting but also raised some concerns. We would like to invite you to submit a revision if you can address the reviewers' key concerns, especially those raised by reviewer 1. The main comment of reviewer 1 is related to the interpretation of the in silico results. Please consider if you can add experimental data to support the method or alternatively revise the text so it will better represent the applicability of the method.
We hope that the comments below will prove constructive, and I would be happy to discuss any plans for a revision once you've had a chance to consider the points raised by the reviewers.
Best Regards,
Sahar
Sahar Melamed, PhD
Recommender at PCI Genomics
You developed a computational pipeline to design 'toehold' riboswitches for specific RNA trigger sequences. The pipeline predicts which toehold switch design adheres to the secondary structure of a known toehold switch, through a combination of RNA secondary structure prediction and subsequent free energy prediction of that structure to assess its stability. A further in silico validation of the method via molecular dynamics is included.
While the method is in itself well designed and likely very helpful in relation to toehold switch design, the interpretation of the in silico results is overly optimistic. Typically, in silico methods are very good at separating what might work from what likely does not work (in this case, a particular toehold switch), but here there is no assessment of how reliable your method is, as validated by experimental data. This is an essential component that is missing in your study: is there (independent) data available on toehold switches that are know to work, and is your approach able to detect/score those? Do you have any independent experimental data that illustrates that this method in fact works well (you mention the iGEM project A.D.N. - did this use your approach, and if so how well did it work)?
In addition, RNA is a notoriously flexible molecule issue, how well can the silico RNA predictions that you are using account for that - do your ‘free energy’ calculations take entropy into account? What could go wrong in these calculations? These issues are not addressed - but should be.
Finally, your statements should always mention ‘predicted’ when this is where your information comes from, e.g. page 10, section 3.2, should say ‘predicted secondary structure’ - as this is what it is, there is no experimental validation.
You should address these issues, or tone down your statements about the real-life applicability of your method, as at the moment it is impossible to assess whether your pipeline works in reality (or not).
I found the preprint clear well written, with only a few typos and formatting issues (listed below). As a non-specialist reviewer, there were two terms that I found difficult to understand and I recommend that the authors include a few words of explanation about each of them in the paper to make it more accessible to a broad readership: "orthogonality" and "overregard". I also found some sentences that sounded quite finalist, and recommend reformulating them to avoid this: "nature has explored many different regulatory mechanisms"; "capable of regulating transcription and translation to optimize the use of resources" (I suggest "capable of regulating transcription and translation, thereby optimizing the use of resources" that does not convey this finalist undertone).
Minor typos and formatting issues:
1) please put line numbers in your resubmitted preprint - it is tricky to provide feedback without them
2) in part 1.1., "RNA molecules, which typically" should be "RNA molecules that typically"
3) "have led to several different designs.-" please remove the unnecessary hyphen
4) "a ribosome binding site" -> "a ribosome-binding site"
5) in panel 3.2 of Figure 2, "cannonical" -> "canonical"
6) in part 3.1, "did not unwind which would lead" -> "did not unwind, which would have lead"
7) in part 4.1, "was stable in the conditions" -> "was stable under the conditions"
8) "with their complementary sequences fluctuates the most often during the simulation" -> "with their complementary sequences fluctuated most often during the simulation"
9) "presented a similar structure to the one" -> "presented a structure similar to the one"
10) "All switches have a negative energy that predicts" -> "All switches had a negative energy that predicted"
11) nearly all references are in "sentence case" except three that are in "Title Case" (refs. 1, 9 and 22), please put them in "sentence case" as well
12) please remove "Chapter one - " from the begining of the title of reference 22, and please remove also the number "1" at the end of the title
13) in the PDF preprint, clicking on the references link to a paperpile URL that gives an error message. Please remove these useless links or replace them with proper DOI links.