

Bacteria and phages have coexisted and coevolved for a long time. Phages are bacteria-infecting viruses, with a symbiotic status sensu lato, meaning they can be pathogenic, commensal or mutualistic. Thus, the association between bacteria phages has probably played a key role in the high adaptability of bacteria to most - if not all – of Earth’s ecosystems, including other living organisms (such as eukaryotes), and also regulate bacterial community size (for instance during bacterial blooms).
As genetic entities, phages are submitted to mutations and natural selection, which changes their DNA sequence. Therefore, comparative genomic analyses of contemporary phages can be useful to understand their evolutionary dynamics. International initiatives such as SEA-PHAGES have started to tackle the issue of history of phage-bacteria interactions and to describe the dynamics of the co-evolution between bacterial hosts and their associated viruses. Indeed, the understanding of this cross-talk has many potential implications in terms of health and agriculture, among others.
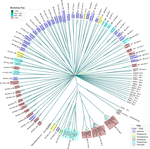
The work of Koert et al. (2021) deals with one of the largest groups of bacteria (Actinobacteria), which are Gram-positive bacteria mainly found in soil and water. Some soil-born Actinobacteria develop filamentous structures reminiscent of the mycelium of eukaryotic fungi. In this study, the authors focused on the Streptomyces clade, a large genus of Actinobacteria colonized by phages known for their high level of genetic diversity.
The authors tested the hypothesis that large exchanges of genetic material occurred between Streptomyces and diverse phages associated with bacterial hosts. Using public datasets, their comparative phylogenomic analyses identified a new cluster among Actinobacteria–infecting phages closely related to phages of Firmicutes. Moreover, the GC content and codon-usage biases of this group of phages of Actinobacteria are similar to those of Firmicutes.
This work demonstrates for the first time the transfer of a bacteriophage lineage from one bacterial phylum to another one. The results presented here suggest that the age of the described transfer is probably recent since several genomic characteristics of the phage are not fully adapted to their new hosts. However, the frequency of such transfer events remains an open question. If frequent, such exchanges would mean that pools of bacteriophages are regularly fueled by genetic material coming from external sources, which would have important implications for the co-evolutionary dynamics of phages and bacteria.
References
Koert, M., López-Pérez, J., Courtney Mattson, C., Caruso, S. and Erill, I. (2021) Evidence for shared ancestry between Actinobacteria and Firmicutes bacteriophages. bioRxiv, 842583, version 5 peer-reviewed and recommended by Peer community in Genomics. doi: https://doi.org/10.1101/842583
DOI or URL of the preprint: https://doi.org/10.1101/842583
Version of the preprint: 2.0
Good morning,
The issues mentioned (ML tree figure, stating how the evolutionary model for ML was selected) have been addressed and a final version is posted to Zenodo and BioRxiv.
https://zenodo.org/record/4392879
https://www.biorxiv.org/content/10.1101/842583v5
Thank you very much,
Steve Caruso
Dear Steven,
Thanks for this new version containing a new ML phylogenetic tree that confirms your previous findings using bioNJ: the close relationship between the phages of some Actinobacteria with those of Lactococcus and Faecalibacterium. I am glad to confirm that your preprint is ready to be recommended by PCI Genomics, pending three minor concerns that would need to be addressed.
Since ML has been proven to perform better than bioNJ for phylogenetic inference, I strongly think that the ML tree should appear as a main figure, while the bioNJ should be included in the supplementary data.
Also, it would be great if you could include a sentence clarifying how you selected the model: did you do a model selection step, or did you select the model without testing based on its properties? If so, why?
Last but not least, please check that the Zenodo link provided in the final pdf is the correct one, and please check also the figure order and captions both in the manuscript and the supplementary material (particularly as two of the figures will change their actual order).
Thanks in advance for your contribution to PCI Genomics and looking forward to reading your very last version!
All the best,
Denis
DOI or URL of the preprint: https://doi.org/10.1101/842583
Version of the preprint: 2.0
Good morning,
The correction has been made as requested. The response/reply and proof are attached. The updated versions have been uploaded to both bioRxiv and Zenodo.
https://www.biorxiv.org/content/10.1101/842583v4
Paper https://zenodo.org/record/4392879
Supporting material https://zenodo.org/record/4584810
Thank you,
Steve Caruso
Dear authors,
I have received the second round of reviews and they emphasize that your preprint has now much improved.
However, there is still a critical issue concerning the method used for phylogenetics analyses.
As a recommender, I agree with the reviewer who asks that you switch to Maximum-Likelihood (ML) phylogenetic trees for your analyses. I hope that you will be able to provide us with these new analyses for a new (and likely final) version of your preprint.
Thanks a lot in advance for your understanding.
I have read the new version of the article and the authors’ responses to the various questions and I think that they did a good job to address them and to include relevant additional information in the manuscript. Therefore, I support the publication of this revised version.
https://doi.org/10.24072/pci.genomics.100008.rev21I’m satisfied with all modifications to the new version of the manuscript. However, I can not understand the persistence of the authors to keep their phylogenetic analyses based on the non-accurate Neighbor-Joining (NJ) method.
I fully understand the difficulty to obtain the Bayesian inference, especially with highly divergent amino acid sequence alignments. Still, the same doesn’t apply to the Maximum-Likelihood method. Nowadays, it’s easy to compute a robust Maximum-Likelihood (ML) phylogenetic tree using a high number of sequences using user-friendly software i.e. IQ-TREE2. IQ-TREE2 not only a fast and easy to use software but also can automatically choose the best-fitting substitution model. Finally, a better Visualization of the phylogenetic trees is highly appreciated (Figures 3 and 4).
Overall, I appropriate the time and effort of the authors to complete this revised version. Then, I recommend the preprint after the authors provide the Maximum-Likelihood (ML) phylogenetic trees.
https://doi.org/10.24072/pci.genomics.100008.rev22DOI or URL of the preprint: https://doi.org/10.1101/842583
Dear authors,
The reviewers did their assessment on your preprint. They all consider that this sharp and focused study on evolution of phages and their bacterial host deserve to be recommended, and I agree too. However, there are – as usual – several discussed issues that need to be fixed. The issue of a possible confusion between host-shift and the so-called « genome relatedness » by Reviewer 3 is to my opinion a critical one. I do hope these assessments will help you to improve your preprint and I am waiting forwards to reading a new revised version of it.
Additional requirements of the managing board:
As indicated in the 'How does it work?’ section and in the code of conduct, please make sure that:
-Data are available to readers, either in the text or through an open data repository such as Zenodo (free), Dryad or some other institutional repository. Data must be reusable, thus metadata or accompanying text must carefully describe the data.
-Details on quantitative analyses (e.g., data treatment and statistical scripts in R, bioinformatic pipeline scripts, etc.) and details concerning simulations (scripts, codes) are available to readers in the text, as appendices, or through an open data repository, such as Zenodo, Dryad or some other institutional repository. The scripts or codes must be carefully described so that they can be reused.
-Details on experimental procedures are available to readers in the text or as appendices.
-Authors have no financial conflict of interest relating to the article. The article must contain a "Conflict of interest disclosure" paragraph before the reference section containing this sentence: "The authors of this preprint declare that they have no financial conflict of interest with the content of this article." If appropriate, this disclosure may be completed by a sentence indicating that some of the authors are PCI recommenders: “XXX is one of the PCI XXX recommenders.”
This brief article discusses the similarity between the genome sequences of several Actinobacteria-infecting phages and those of a small group of Firmicutes phages. The authors hypothesize that this similarity suggests that these Actinobacteria phages have shifted from an ancestor infecting Firmicutes towards their current actinobacterial hosts. This host shift may have occurred recently, as supported by the GC content of these phages which is significantly different from that of their hosts, indicating that they are still in the process of ameliorating it to adapt to their new hosts.
The article is well written, concise and clear. The results are interesting and the conclusions seem to be well supported. I only have two comments:
1- Protein sequence phylogenetic analysis: The authors study the phylogeny of terminase protein sequences using the Neighbor-Joining (NJ) method with the JTT substitution model. It is not mentioned if all positions in the multiple sequence alignment were used for phylogenetic inference or if the alignment was trimmed (e.g., to remove gaps and/or ambiguously aligned positions). This needs to be clarified. In addition, NJ with the JTT model is probably not the best approach to analyze fast-evolving sequences as those frequently found in viruses. The authors should instead use maximum likelihood and/or Bayesian inference with a more appropriate substitution model. Programs like IQ-tree offer the possibility to choose the best-fitting model automatically.
2- The GC content alone remains a very rough estimate of adaptation to the host. It would be useful to have data about more precise indicators, such as the Codon Adaptation Index (CAI). See for example https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4032129/
https://doi.org/10.24072/pci.genomics.100008.rev12This work reports on the relatedness of a cluster of phages infecting Firmicutes and Actinobacteria. It shows several genomic alignments and trees, and terminates by a GC% analysis. Data are convincing but need to be presented more rigorously. Below a list of changes that are necessary prior acceptance, in my view.
Major points
There is confusion in the mind of authors between genome relatedness and host range. One should keep in mind that a phage ancestor of the described cluster might have infected the last common ancestor of the Actinobacteria and Firmicutes branches, and then diverged along the various branches. There is no need to invoke wide host range when one finds genome relatedness. Or it should be tested experimentally. The true and convincing cases of large host range concern identical phages (meaning 99 to 100% identity) infecting different genera/families (e. g. https://www.ncbi.nlm.nih.gov/pubmed/29615108). This same reference should be added when FPoengus is mentioned. It also reports the relatedness of FPoengus with the Lactococcus phage 1706. The first sentence of the abstract is particularly obscure. Phages are known NOT to display broad host ranges. May be a negation was lost along the line? But the discussion comes back with similar confusions: “it is well known that phages will often infect several hosts within the same genus”: no reference here. In fact the opposite is known, as most phages infect a few strains per species. Ref 28 on Mu is given to assert that phage systems have been “engineered” to transcend genus boundaries. In the case of Mu, it is a natural trait, not man made. The ref 29 about “virus transfer across phyla” does not show phage infection, but (putative) phage transduction.
Figures are difficult to read. In Figure 1 (where phage genomes are), scripts are too small. Genes have no arrow (a big weakness of phamerator maps). Color codes are not explained. Shaded area have no scale. Figure 5 is really difficult to read, with all these bars crossing each other. It should be possible (with prism for instance) to display all points as clouds, with genera on x axis, and GC% on y axis. A color code would permit to contrast bacterial genomes and phage clusters. One would see how many members are being compared in each case. The difference between B1 and other Streptomyces clusters is not clear.
Figure legends should be greatly improved. It is scarcely understandable which data were used to build each of the trees. Fig5 legend is obscure. The term ‘cluster’ is repeated everywhere in this legend. Which Faecalibacterium species were tested, how was the representative member chosen ? Actually, this species might be a genus (https://www.ncbi.nlm.nih.gov/pubmed/30547746).
Annotation of the genes common to this phage cluster should go a bit deeper. In particular, gp58 of RavenPuff is annotated as putative RNA polymerase in oengus. The Koonin group recently studied a group of phages with genes distantly related to RNA polymerases (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5736458/). A further search in this direction would provide more strength to this study. Could this gene be considered a marker gene of the cluster?
Low et al. published a remarkable and remarked concatenated tree of phage genomes last year (https://www.ncbi.nlm.nih.gov/pubmed/31110365). Where is the phage cluster in this tree ? Does it group well? Please situate your work relative to Low et al. Could you try building up a tree with your dataset following the footsteps of this paper?
Sullivan’s group has proposed a network clustering v-CONTACT approach (https://www.ncbi.nlm.nih.gov/pubmed/31061483) which works well at the genus level apparently. How do your genomes cluster in such graphs?
The two above-mentioned papers are seminal in the field of phage genomics, they should be referred to, and put into dialog with your work.
Minor points
First paragraph of results, just before ref 21, ‘consistent with that observed for other Siphoviridae’ is too vague, you probably mean consistent with Cluster J of Mycophages (this is ref21 title)?
Second Result paragraph: clusters AM AU and AW were not introduced: what are they referring to, how were they built? For a broad readership, insert “Actinobacteria hosts such as” just after “elicited significant hits against…”.
Same paragraph: Why is ref 5 mentioned for the phage 1706?
End of last paragraph: Figure SX is probably S2.
Figure 3 and 4: make broader lines, one hardly sees their colors.
Discussion: the conservation of two large genomic blocks does not suggest to me that they are ‘primary functional elements’, they are just less often exchanged. They can constitute the backbone of this cluster if you want. But in phage genomes one hardly thinks in terms of essential and accessory genes.
I am not sure that the closer relatedness in GC% is a proof of ancestry. Compared to the wealth of Lactococcus phages sequenced, this 1706 is really an outsider. In Faecalibacterium, much less is known, but FP_oengus was an orphan in the Cornuault et al study (https://www.ncbi.nlm.nih.gov/pubmed/29615108 ).
I hope authors understood they should revise their mind on the “rapid jump over order and phylum” (sentence just after ref 5 at the end of the discussion) as mentioned as my top criticism on this paper. Or they should prove this jump experimentally. It would be great.